
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,533,282 – 16,533,456 |
| Length | 174 |
| Max. P | 0.911971 |

| Location | 16,533,282 – 16,533,396 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -16.31 |
| Energy contribution | -15.03 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16533282 114 - 23771897 UAGGGGAGUGCCCGCCCAAGAGGGGGGGGGGUGCGGUGUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGAAAAUAUCCAAAGUAUCUGUAGCGCUAGAAAAGUGCAAGCAAUCCG ....(((.(((((.(((....))).)).(((((((((((.((((((........)))))).)))))(((.....)))...))))))...(((((.....)))))..))).))). ( -42.00) >DroSec_CAF1 146521 108 - 1 UAGGGGAGUGCCCGCCCAAAAGGGGACGGGG------GUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGAAAAUAUCCAAAGUAUCUGUAGCGCUAGAAAAGUGCAAGCAAUCCG ..(((((((.(((((((.....))).)))).------).))).((((.(((.....)))...))))(((.....)))............(((((.....)))))......))). ( -33.60) >DroSim_CAF1 149623 108 - 1 UAGGGGAGUGCCCGCCCAAAGGGGGACUUGG------GUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGAAAAUAUCCAAAGUAUCUGUAGCGCUAGAAAAGUGCAAGCAAUCCG ....(((.(((..(((((..((....)))))------))..((((((.(((.....))).......(((.....)))....))))))..(((((.....)))))..))).))). ( -32.60) >DroEre_CAF1 144335 99 - 1 UAGGGGAGUGCCC----AA-GGAG----GAG------GUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGGAAAUAUCUGAAGUAUCUGUAGCGCUAGAGAAGUGCAACCAAUCCG ..(((.....)))----..-(((.----..(------((..((((((.(((((..(((((....)))))....)))))...))))))..(((((.....))))).)))..))). ( -30.90) >DroYak_CAF1 147403 99 - 1 UAGGGGAGUGCCC----AA-GGAG----GGG------GUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGGAAAUAUCUGAAGUAUCUGUAGCGCUAGAAAAGUGCAACCAAUCCG ..(((.....)))----..-(((.----.((------....((((((.(((((..(((((....)))))....)))))...))))))..(((((.....)))))..))..))). ( -32.70) >DroAna_CAF1 147746 86 - 1 UAGGGGAGUGUCC----AA-GGAG----GCG------GAGGUGGAGAUGGGAUCGACUCAGAUCAUGACACACA-------------GAUCGCUGGAAAAGUGCAACCAAUCCC ...((((((.((.----..-.)).----))(------(..(((.......((((......))))......))).-------------...((((.....))))...))..)))) ( -21.72) >consensus UAGGGGAGUGCCC____AA_GGAG____GGG______GUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGAAAAUAUCCAAAGUAUCUGUAGCGCUAGAAAAGUGCAACCAAUCCG ..(((.....))).......(((.................((.((((.(((.....)))...)))).))....................(((((.....)))))......))). (-16.31 = -15.03 + -1.27)
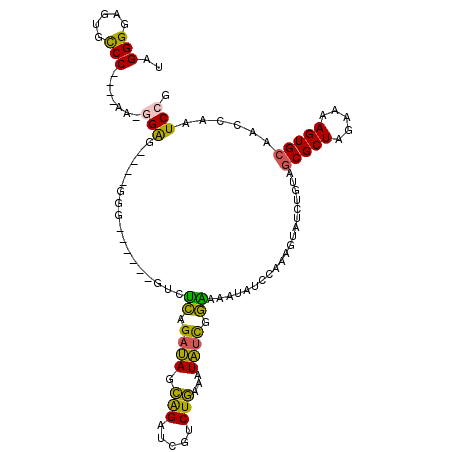

| Location | 16,533,322 – 16,533,421 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -18.54 |
| Energy contribution | -17.18 |
| Covariance contribution | -1.36 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16533322 99 - 23771897 AACAAAGGAGGGGCAUUCAAUUGAUUAGGGGAGUGCCCGCCCAAGAGGGGGGGGGGUGCGGUGUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGAAAAUA .........(((((((((....))........(..(((.(((.......))).)))..).))))))).((((.(((.....)))...))))........ ( -30.70) >DroSec_CAF1 146561 93 - 1 AACAAAGGAGGGGCAUUCAAUUGAUUAGGGGAGUGCCCGCCCAAAAGGGGACGGGG------GUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGAAAAUA .......(((((((((((..(......)..))))))))((((....(....)..))------))))).((((.(((.....)))...))))........ ( -30.80) >DroSim_CAF1 149663 93 - 1 AACAAAGGAGGGGCAUUCAAUUGAUUAGGGGAGUGCCCGCCCAAAGGGGGACUUGG------GUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGAAAAUA .......(((((((((((..(......)..))))))))(((((..((....)))))------))))).((((.(((.....)))...))))........ ( -32.00) >DroEre_CAF1 144375 84 - 1 AACAAAGGAGGGGCAUUCAAUUGAUUAGGGGAGUGCCC----AA-GGAG----GAG------GUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGGAAAUA ..........((((((((..(......)..))))))))----..-....----...------.((((.((((.(((.....)))...)))))))).... ( -23.00) >DroYak_CAF1 147443 84 - 1 CACAAAGGAGGGGCAUUCAAUUGAUUAGGGGAGUGCCC----AA-GGAG----GGG------GUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGGAAAUA ..........((((((((..(......)..))))))))----..-....----...------.((((.((((.(((.....)))...)))))))).... ( -23.00) >DroAna_CAF1 147773 84 - 1 AACAAAGGCGGCGCAUUCAAUUGAUUAGGGGAGUGUCC----AA-GGAG----GCG------GAGGUGGAGAUGGGAUCGACUCAGAUCAUGACACACA .........((.((((((..(......)..))))))))----..-....----...------...(((.......((((......))))......))). ( -15.02) >consensus AACAAAGGAGGGGCAUUCAAUUGAUUAGGGGAGUGCCC____AA_GGAG____GGG______GUCUCAGAUAGCAGAUCGUCUGAAAUAUCGGAAAAUA ..........((((((((..(......)..))))))))...........................((.((((.(((.....)))...)))).))..... (-18.54 = -17.18 + -1.36)
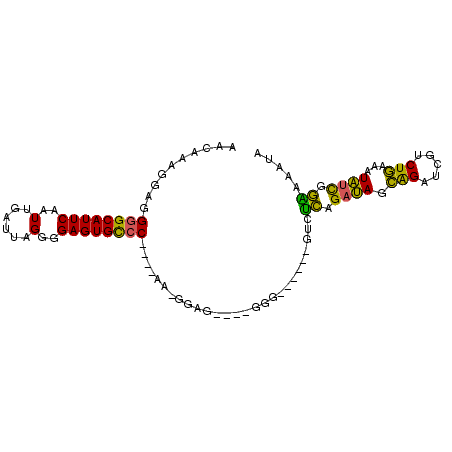

| Location | 16,533,362 – 16,533,456 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 78.53 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -11.74 |
| Energy contribution | -11.30 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.765975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16533362 94 - 23771897 -AAUGCCGAUGGGAAUGGGAAUGGG---UUGAAAAUGCAAACAAAGGAGGGGCAUUCAAUUGAUUAGGGGAGUGCCCGCCCAAGAGGGGGGGGGGUGC -....((.((....)).))(((.((---((((..((((...(......)..)))))))))).)))......(..(((.(((.......))).)))..) ( -27.10) >DroSec_CAF1 146598 91 - 1 -AAUGCCGAUGCGAAUGGGAAUGGG---UUGAAAAUGCAAACAAAGGAGGGGCAUUCAAUUGAUUAGGGGAGUGCCCGCCCAAAAGGGGACGGGG--- -....(((.(.(...((((..((..---(((......))).))......((((((((..(......)..)))))))).))))...).)..)))..--- ( -24.20) >DroSim_CAF1 149700 91 - 1 -AAUGCCGAUGCGAAUGGGAAUGGG---UUGAAAAUGCAAACAAAGGAGGGGCAUUCAAUUGAUUAGGGGAGUGCCCGCCCAAAGGGGGACUUGG--- -....((.((....)).))..((((---(((......))..........((((((((..(......)..))))))))))))).(((....)))..--- ( -24.70) >DroYak_CAF1 147480 80 - 1 -AAUGCCGAUGCGAU--GGCAUGGG---UUGCAAAUGCACACAAAGGAGGGGCAUUCAAUUGAUUAGGGGAGUGCCC----AA-GGAG----GGG--- -...(((.((((...--.)))).))---)(((....)))..........((((((((..(......)..))))))))----..-....----...--- ( -24.70) >DroAna_CAF1 147810 79 - 1 AAAUGCCUUUGG-------AGGGGGCUUUGGAGAAUGCAAACAAAGGCGGCGCAUUCAAUUGAUUAGGGGAGUGUCC----AA-GGAG----GCG--- ....((((((..-------....(.(((((...........))))).)((.((((((..(......)..))))))))----..-))))----)).--- ( -21.20) >consensus _AAUGCCGAUGCGAAUGGGAAUGGG___UUGAAAAUGCAAACAAAGGAGGGGCAUUCAAUUGAUUAGGGGAGUGCCCGCCCAA_GGGGG___GGG___ .....((.((..........)).))........................((((((((..(......)..))))))))..................... (-11.74 = -11.30 + -0.44)

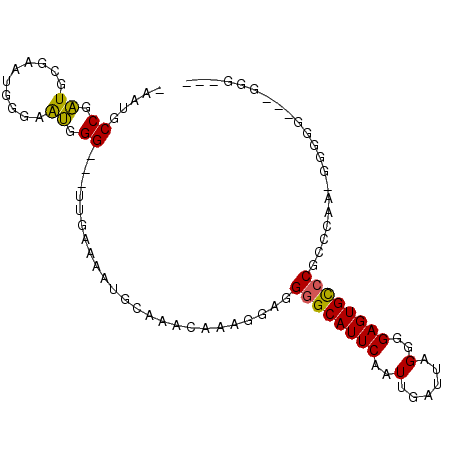
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:04 2006