
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,522,106 – 16,522,230 |
| Length | 124 |
| Max. P | 0.998779 |

| Location | 16,522,106 – 16,522,202 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 95.62 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
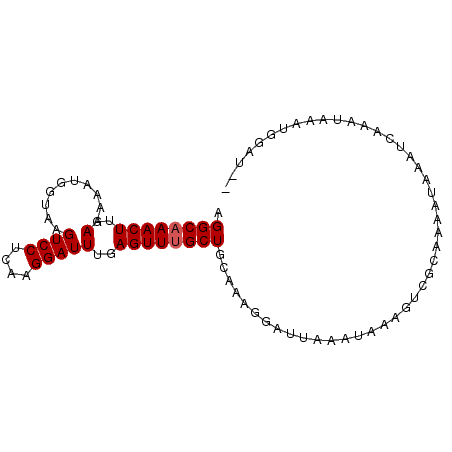
>3L_DroMel_CAF1 16522106 96 + 23771897 UUCAAUUGCAAGACGCAAAUCGUAAACCCGAAAAGGCAAACUUGAAAAAUGGUAAGUCCUCAAGGAUUUGAGUUUGCUGCAAAGGAUUAAGUAAAG .((..((((..........(((......)))...((((((((..(..........((((....)))))..))))))))))))..)).......... ( -20.60) >DroSec_CAF1 135668 96 + 1 UUCAAUUGCAAGACGCAAAUCGUAAACCCGAAAAGGCAAACUUGAAAAAUGGUAAGUCCUCAAGGAUUUGAGUUUGCUGCAAAGGAUUAAAUAAAG .((..((((..........(((......)))...((((((((..(..........((((....)))))..))))))))))))..)).......... ( -20.60) >DroSim_CAF1 138619 96 + 1 UUCAAUUGCAAGACGCAAAUCGUAAACCCGAAAAGGCAAACUUGAAAAAUGGUAAGUCCUCAAGGAUUUGAGUUUGCUGCAAAGGAUUAAAUAAAG .((..((((..........(((......)))...((((((((..(..........((((....)))))..))))))))))))..)).......... ( -20.60) >DroEre_CAF1 131325 94 + 1 UUCAAUUGCAAGACGCAAAUCGUAAACCCGAAAAGGCAAACUUGAAAAAUGGUAAGUCCUCUUGGAUUUGAGUUUGCUGCA-AGGAUUA-UUGAUG .((((((((.....)))).........((.....((((((((..(..........((((....)))))..))))))))...-.))....-)))).. ( -21.90) >DroYak_CAF1 135882 95 + 1 UUCAAUUGCAAGACGCAAAUCGUAAACCCGAAAAGGCAAACUUGAAAAAUGGUAAGUCCUCUUGGAUUUGAGUUGGCUGCAAAGGAUUA-UUAAAG .((..(((((.(((((...(((......)))....))...............(((((((....))))))).)))...)))))..))...-...... ( -18.00) >consensus UUCAAUUGCAAGACGCAAAUCGUAAACCCGAAAAGGCAAACUUGAAAAAUGGUAAGUCCUCAAGGAUUUGAGUUUGCUGCAAAGGAUUAAAUAAAG .....((((.....)))).........((.....((((((((..(..........((((....)))))..)))))))).....))........... (-19.82 = -20.02 + 0.20)

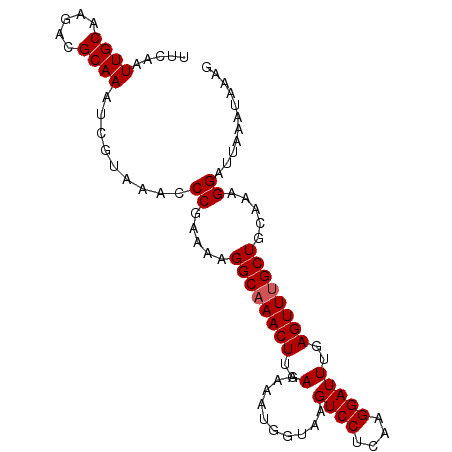
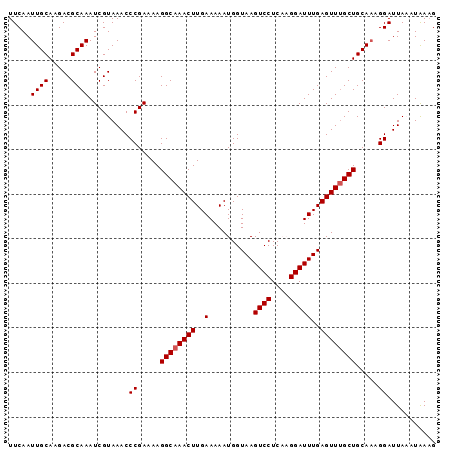
| Location | 16,522,106 – 16,522,202 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 95.62 |
| Mean single sequence MFE | -20.62 |
| Consensus MFE | -18.82 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16522106 96 - 23771897 CUUUACUUAAUCCUUUGCAGCAAACUCAAAUCCUUGAGGACUUACCAUUUUUCAAGUUUGCCUUUUCGGGUUUACGAUUUGCGUCUUGCAAUUGAA ..........((..((((((...((.((((((.(((((((........)))))))((..((((....))))..)))))))).)).))))))..)). ( -21.30) >DroSec_CAF1 135668 96 - 1 CUUUAUUUAAUCCUUUGCAGCAAACUCAAAUCCUUGAGGACUUACCAUUUUUCAAGUUUGCCUUUUCGGGUUUACGAUUUGCGUCUUGCAAUUGAA ..........((..((((((...((.((((((.(((((((........)))))))((..((((....))))..)))))))).)).))))))..)). ( -21.30) >DroSim_CAF1 138619 96 - 1 CUUUAUUUAAUCCUUUGCAGCAAACUCAAAUCCUUGAGGACUUACCAUUUUUCAAGUUUGCCUUUUCGGGUUUACGAUUUGCGUCUUGCAAUUGAA ..........((..((((((...((.((((((.(((((((........)))))))((..((((....))))..)))))))).)).))))))..)). ( -21.30) >DroEre_CAF1 131325 94 - 1 CAUCAA-UAAUCCU-UGCAGCAAACUCAAAUCCAAGAGGACUUACCAUUUUUCAAGUUUGCCUUUUCGGGUUUACGAUUUGCGUCUUGCAAUUGAA ..((((-(......-(((((((((.((((((((((((((.(..((..........))..))))))).))))))..)))))))....))))))))). ( -19.40) >DroYak_CAF1 135882 95 - 1 CUUUAA-UAAUCCUUUGCAGCCAACUCAAAUCCAAGAGGACUUACCAUUUUUCAAGUUUGCCUUUUCGGGUUUACGAUUUGCGUCUUGCAAUUGAA ......-...((..((((((...((.((((((...(((((........)))))..((..((((....))))..)))))))).)).))))))..)). ( -19.80) >consensus CUUUAAUUAAUCCUUUGCAGCAAACUCAAAUCCUUGAGGACUUACCAUUUUUCAAGUUUGCCUUUUCGGGUUUACGAUUUGCGUCUUGCAAUUGAA ..........((..((((((...((.((((((...(((((........)))))..((..((((....))))..)))))))).)).))))))..)). (-18.82 = -19.02 + 0.20)

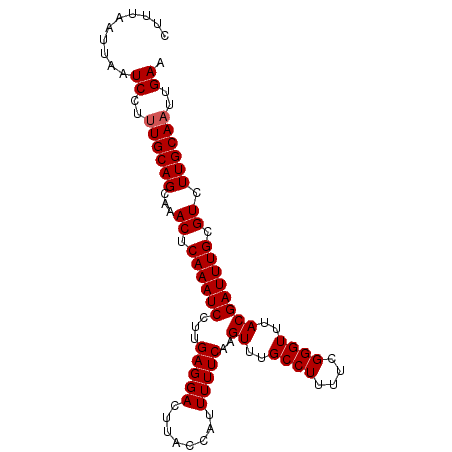
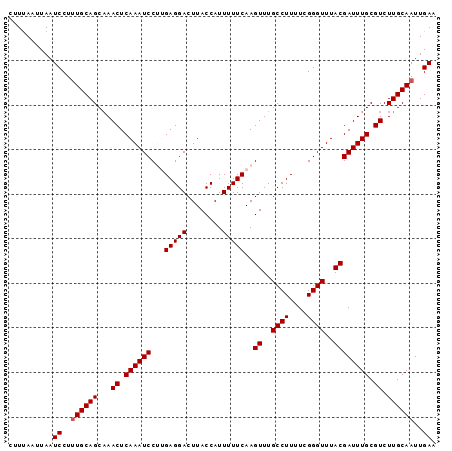
| Location | 16,522,139 – 16,522,230 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 87.26 |
| Mean single sequence MFE | -15.56 |
| Consensus MFE | -13.72 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16522139 91 + 23771897 AGGCAAACUUGAAAAAUGGUAAGUCCUCAAGGAUUUGAGUUUGCUGCAAAGGAUUAAGUAAAGUCGCUAAACAAAUCAAAUAAAUUGAUCA .((((((((..(..........((((....)))))..))))))))....((((((......)))).))......(((((.....))))).. ( -17.10) >DroSec_CAF1 135701 89 + 1 AGGCAAACUUGAAAAAUGGUAAGUCCUCAAGGAUUUGAGUUUGCUGCAAAGGAUUAAAUAAAGUCGCAAAAUAAAUCAAAUAAAUGGAU-- .((((((((..(..........((((....)))))..))))))))......((((......))))........................-- ( -15.60) >DroSim_CAF1 138652 89 + 1 AGGCAAACUUGAAAAAUGGUAAGUCCUCAAGGAUUUGAGUUUGCUGCAAAGGAUUAAAUAAAGUCGCAAAAUAAAUCAAAUAAAUGGAU-- .((((((((..(..........((((....)))))..))))))))......((((......))))........................-- ( -15.60) >DroEre_CAF1 131358 81 + 1 AGGCAAACUUGAAAAAUGGUAAGUCCUCUUGGAUUUGAGUUUGCUGCA-AGGAUUA-UUGAUGUCGGAUAAUAAAUCAAAGAA-------- .((((((((..(..........((((....)))))..))))))))...-.......-(((((............)))))....-------- ( -16.50) >DroYak_CAF1 135915 90 + 1 AGGCAAACUUGAAAAAUGGUAAGUCCUCUUGGAUUUGAGUUGGCUGCAAAGGAUUA-UUAAAGGCGAAAAAUCAAUCAAAUAAAUUGAUAU .(((.((((..(..........((((....)))))..)))).)))...........-.............((((((.......)))))).. ( -13.00) >consensus AGGCAAACUUGAAAAAUGGUAAGUCCUCAAGGAUUUGAGUUUGCUGCAAAGGAUUAAAUAAAGUCGCAAAAUAAAUCAAAUAAAUGGAU__ .((((((((..(..........((((....)))))..)))))))).............................................. (-13.72 = -13.92 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:55 2006