
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,511,918 – 16,512,054 |
| Length | 136 |
| Max. P | 0.826309 |

| Location | 16,511,918 – 16,512,026 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -14.62 |
| Consensus MFE | -8.74 |
| Energy contribution | -9.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
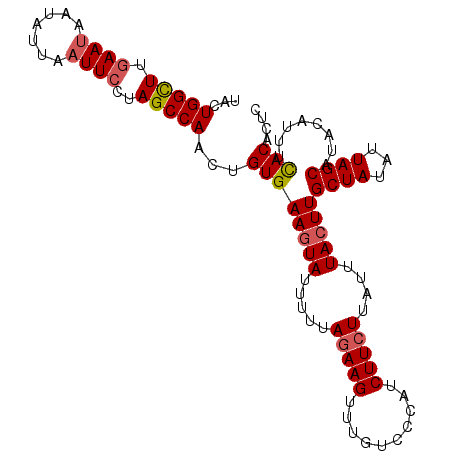
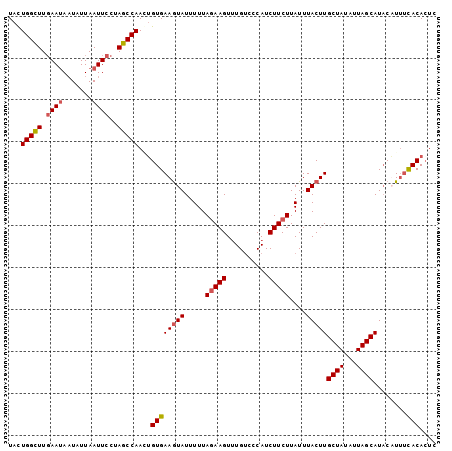
>3L_DroMel_CAF1 16511918 108 - 23771897 CCAAGUGUGAAGUAUUUUUAGAAGUUUGUCCCAUCUUCUUAUUUACUUGCUAUAUUAGCAUACUUUUUACAAUCUU-AUCGGCAUAUGUGCCAAAUUAUUUUAUACUCU ...(((((((((((.(((.(((((..........)))))........(((((...)))))................-...((((....))))))).))))))))))).. ( -19.70) >DroSec_CAF1 117974 92 - 1 CCAACUGUGAAGUAUUUUUAGAAGUUUGUCCCAUCUUCUUAUUUACUUGCUAUAUUAGCAUACAUUUCACACUCUU-----------------AAUUAUUCAAUACUCU .....(((((((((.....(((((..........)))))....))).(((((...))))).....)))))).....-----------------................ ( -13.20) >DroSim_CAF1 127945 92 - 1 CCAACUGUGAAGUAUUUUUAGAAGUAUGUCCCAUCUUCUUAUUUACUUGCUAUAUUAGCAUACAUUUCACACUCUA-----------------AGUUAUUCAAUACUCU .....(((((((((.....(((((.(((...))))))))....))).(((((...))))).....)))))).....-----------------................ ( -14.30) >DroYak_CAF1 125116 109 - 1 CCAAAGGUGAAAUAUUUUUAAAAGUUUGCCCCAUCUUCUUAUUUACUUGCUAUUUUAGCAUUCUUGACACACUCUUUAUCGGCAAAUGAGUCAAAUUAUUAAAUAUUCU ..........((((.(((.....(((((((.................(((((...))))).....((.....))......))))))).....))).))))......... ( -11.30) >consensus CCAACUGUGAAGUAUUUUUAGAAGUUUGUCCCAUCUUCUUAUUUACUUGCUAUAUUAGCAUACAUUUCACACUCUU_________________AAUUAUUCAAUACUCU ......((((((((.....(((((..........)))))....)))))((((...))))........)))....................................... ( -8.74 = -9.05 + 0.31)



| Location | 16,511,952 – 16,512,054 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -13.66 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16511952 102 - 23771897 UACUGGUUUGAAUAAUAUUAAUUCCUAGCCAAGUGUGAAGUAUUUUUAGAAGUUUGUCCCAUCUUCUUAUUUACUUGCUAUAUUAGCAUACUUUUUACAAUC ...(((((.((((.......))))..))))).(((.((((((.....(((((..........)))))........(((((...))))))))))).))).... ( -18.10) >DroSec_CAF1 117992 102 - 1 UACUGGCUUGAAUAAUAUUAAUUCCUAGCCAACUGUGAAGUAUUUUUAGAAGUUUGUCCCAUCUUCUUAUUUACUUGCUAUAUUAGCAUACAUUUCACACUC ...(((((.((((.......))))..)))))..(((((((((.....(((((..........)))))....))).(((((...))))).....))))))... ( -21.40) >DroSim_CAF1 127963 102 - 1 UACUGGCUUGAAUAAUAUUAAUUCCUAGCCAACUGUGAAGUAUUUUUAGAAGUAUGUCCCAUCUUCUUAUUUACUUGCUAUAUUAGCAUACAUUUCACACUC ...(((((.((((.......))))..)))))..(((((((((.....(((((.(((...))))))))....))).(((((...))))).....))))))... ( -22.50) >DroYak_CAF1 125151 102 - 1 UAUUGGUUUUAAGAUUAUUAAUUCUUAGCCAAAGGUGAAAUAUUUUUAAAAGUUUGCCCCAUCUUCUUAUUUACUUGCUAUUUUAGCAUUCUUGACACACUC ..(((((..(((((........)))))))))).((..((...((.....)).))..)).................(((((...))))).............. ( -13.60) >consensus UACUGGCUUGAAUAAUAUUAAUUCCUAGCCAACUGUGAAGUAUUUUUAGAAGUUUGUCCCAUCUUCUUAUUUACUUGCUAUAUUAGCAUACAUUUCACACUC ...(((((.((((.......))))..)))))...((((((((.....(((((..........)))))....)))))((((...))))........))).... (-13.66 = -14.22 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:50 2006