| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,839,410 – 1,839,523 |
| Length | 113 |
| Max. P | 0.625140 |

| Location | 1,839,410 – 1,839,523 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.33 |
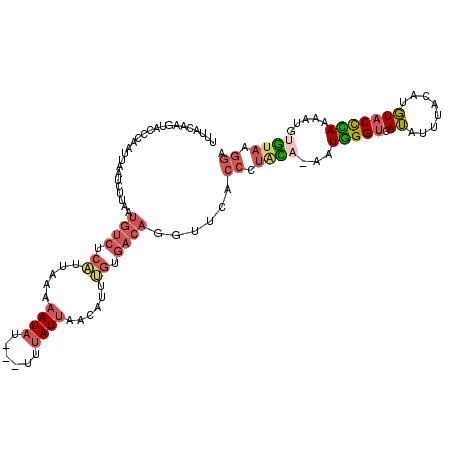
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -6.02 |
| Energy contribution | -7.00 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.27 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
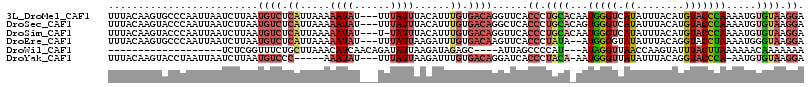
>3L_DroMel_CAF1 1839410 113 - 23771897 UUUACAAGUGCCCAAUUAAUCUUAAUGUCUCAUUAAAAAUAU---UUUAUUUACAUUUGUGACAGGUUCACCCUGCACAAUGGGUCAUAUUUACAUGUACCCAAAAUGUGUAAGGA ...........((........((((((...))))))......---.....(((((((((((.((((.....)))))))))((((((((......))).)))))....)))))))). ( -24.90) >DroSec_CAF1 15229 113 - 1 UUUACAAGUACCCAAUUAAUCUUAAUGUCUCAUUAAAAAUAU---UUUAUUUACAUUUGUGACAGGCUCACCCUGCACAGUGGGUCAUAUUUACAUGUACCCAAAAUGUGUAAGGA .((((((((............((((((...))))))(((((.---..)))))..))))))))........((.((((((.((((((((......))).)))))...)))))).)). ( -24.50) >DroSim_CAF1 16013 112 - 1 UUUACAAGUACCCAAUUAAUCUUAAUGUCUCAUUAAAAAUAU---U-UAUUUACAUUUGUGACAGGUUCACCCUGCACAAUGGGUCAUAUUUACAUGUACCCAAAAUGUGUAAGGA ...........((........((((((...))))))......---.-...(((((((((((.((((.....)))))))))((((((((......))).)))))....)))))))). ( -24.90) >DroEre_CAF1 15032 111 - 1 UUUACAAGUGCCCAAUUAAUCUUAAUGUCUCAUUAAAAAUAU---UUUAUUAAGAUUUGUGACAAGUUCACCCUAUA--AUGGGGUAUAUUUACAGGUACCUAAAAUGGGUAAGGA ....(...((((((...((((((((((...............---..))))))))))(((((...((..(((((...--..)))))..)))))))...........))))))..). ( -22.73) >DroWil_CAF1 22631 90 - 1 -------------------UCUCGGUUUCUGCUUAAACAUCAACAGAUAUUAAGAUAGAGC----AUUAGCCCCAU---AUAGGUUAACCAAGUAUUUACUUAAAAAACAAAAAAA -------------------....(((((((((((((..(((....))).))))).))))).----..(((((....---...))))))))((((....)))).............. ( -12.60) >DroYak_CAF1 15769 106 - 1 UUUACAAGUACCUAAUUAAUCUUAAUGUCCC-----AAAUAU---UUUAUUAAGAUUUGUGACAGGAUCACCCUACA-AAUGGGUUAUAUUUACAGGUACCCA-AAUGUGUAAGGA ((((((.((((((....((((((((((....-----......---..)))))))))).((((....((.((((....-...)))).))..))))))))))...-....)))))).. ( -22.60) >consensus UUUACAAGUACCCAAUUAAUCUUAAUGUCUCAUUAAAAAUAU___UUUAUUAACAUUUGUGACAGGUUCACCCUACA_AAUGGGUCAUAUUUACAUGUACCCAAAAUGUGUAAGGA .........................((((.((.....((((......))))......)).))))......((.((((...(((((.((........))))))).....)))).)). ( -6.02 = -7.00 + 0.98)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:59 2006