
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,496,997 – 16,497,099 |
| Length | 102 |
| Max. P | 0.775864 |

| Location | 16,496,997 – 16,497,099 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -10.24 |
| Energy contribution | -10.35 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16496997 102 + 23771897 GGCCAACGGCAUAAAUAUGUCAAAAACACAACGCUCCGCGUGCUAUAAACAAAUUGUU-UUGCUGUUGCCAAAAAA---AU--AUAUGA-AAUAUUUCAGCAAAAAAAA (((.(((((((.(((((.............((((...)))).............))))-)))))))))))......---..--...(((-(....)))).......... ( -19.97) >DroPse_CAF1 123773 101 + 1 GGCCACCGGCAUAAAUAUGUCAAAAACACAACGCGCCGCGUGCUAUAAACAAAUUGUU-UUGUUGUAGCCAAA-AA---CGGGAGAAAAAAAAAUUGGAGGAAA---GA ..((.(((((((....)))))............(.((.((((((((((.((((....)-))))))))))....-.)---)))).)...........)).))...---.. ( -20.90) >DroWil_CAF1 116953 89 + 1 GGCCACCGGCAUAAAUAUGUCAAAAACACAAUGCGCC--GUGCUAUAAACAAAUUGUU-UUG---UUGCCAAA-AA---AU--AUAAAA-AACAC---A-CAAA---AC ((((((.(((.......(((.....)))......)))--))).....((((((....)-)))---)))))...-..---..--......-.....---.-....---.. ( -12.82) >DroYak_CAF1 109351 98 + 1 GGCCAGCGGCAUAAAUAUGUCAAAAACACAGCGCGCCGCGUGCUGUAAACAAAUUGUU-UUGCUGUUGCCAAA-AA---AU--AUAUGA-AAUAUUUCACCAAA---AA (((.(((((((.(((((..........((((((((...))))))))........))))-)))))))))))...-..---..--...(((-(....)))).....---.. ( -26.07) >DroAna_CAF1 113705 105 + 1 GGCCACCGGCAUAAAUAUGUCAAAAGCACAACGCGCUUCGUUAUAUAAACAAAUUGUUUUUGCCGCUGCCAAA-ACAACAA--AAAUAA-AAAUUUUCCAAAAACAAAA (((...(((((.(((((......((((.......)))).(((.....)))....))))).)))))..)))...-.......--......-................... ( -17.60) >DroPer_CAF1 126928 100 + 1 GGCCACCGGCAUAAAUAUGUCAAAAACACAACGCGCGGCGUGCUAUAAACAAAUUGUU-UUGUUGUAGCCAAA-AA---CGGGAGAAAA-AAAAUUGGAGGAAA---GA ..((.(((((((....)))))..........(.(.((....(((((((.((((....)-))))))))))....-..---)).).)....-......)).))...---.. ( -20.00) >consensus GGCCACCGGCAUAAAUAUGUCAAAAACACAACGCGCCGCGUGCUAUAAACAAAUUGUU_UUGCUGUUGCCAAA_AA___AU__AUAAAA_AAAAUUUCAGCAAA___AA (((...(((((..((((.............(((.....))).............))))..)))))..)))....................................... (-10.24 = -10.35 + 0.11)

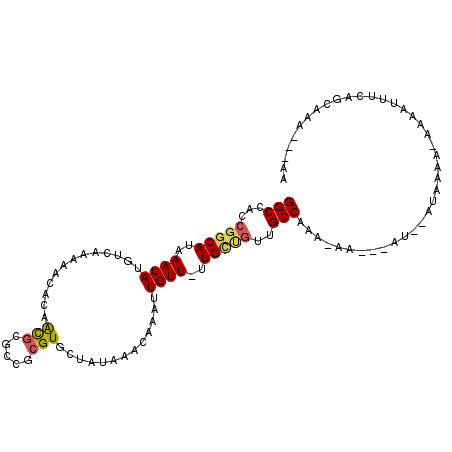
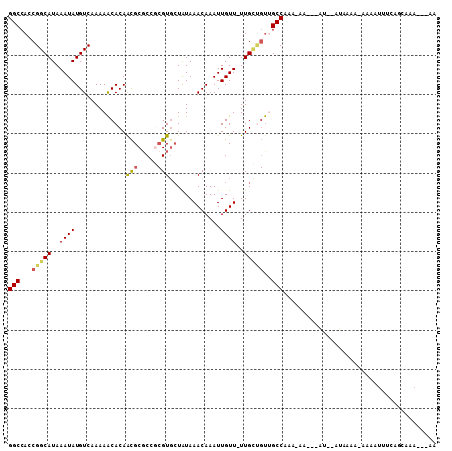
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:46 2006