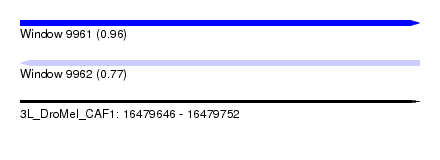
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,479,646 – 16,479,752 |
| Length | 106 |
| Max. P | 0.957878 |

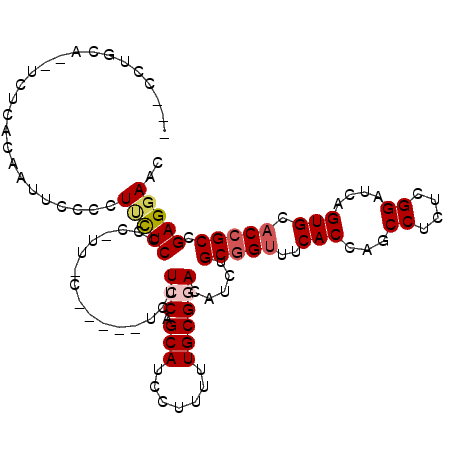
| Location | 16,479,646 – 16,479,752 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -23.71 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16479646 106 + 23771897 AUUCCUCGGCGGUGCACUGAUCCGAGAGGCUGGUGAAACCGCAGAUGUCCGCAAAAAGGAUGCUGGAAAA-----GGGAAGAGGAAUAGGGGAAUUGUAAGC--UGCAGGACC (((((((.(((((.((((...((....))..))))..))))).....((((((.......))).)))...-----.....)))))))..((...((((....--.))))..)) ( -32.80) >DroPse_CAF1 111386 102 + 1 GUUCCUCGGCGGUGCACUGAUCCGAGAGGCUGGUGAAACCGCAAAUGUCCGCAAAGAGGAUGCUGCAAGG-----GAAACGGGGAGGAG----CUUGAGAGACAGGCAGGA-- .((((((((((((.((((...((....))..))))..)))))....((((.......)))).........-----....)))))))..(----((((.....)))))....-- ( -35.20) >DroEre_CAF1 93551 91 + 1 GUUCCUCGGCGGUGCACUGAUCCGAGAGGCUGGUGAAACCGCAGAUGUCCGCAAAAAGGAUGCUGGAAAU------------GGAAAAGGGGAAUUGUGAGC--U-------- (((((((.(((((.((((...((....))..))))..))))).....((((((.......))).)))...------------......))))))).......--.-------- ( -29.00) >DroYak_CAF1 96944 91 + 1 GUUCCUCGGCGGUGCACUGAUCCGAGAGGCUGGUGAAACCGCAGAUGUCCGCAAAAAGGAUGCUGGAAGA------------GGAAAAAAGGAAUUGUGAGU--U-------- .((((((.(((((.((((...((....))..))))..))))).....((((((.......))).))).))------------))))................--.-------- ( -29.40) >DroAna_CAF1 102835 108 + 1 ACUCCUCCGCCGUGCACUGAUCCGAGAGGCUGGUGAAGCCGCAAAUGUCCGCAAACAGAAUGCUGGAAAACACACAGAAAGGGGAGAAGGUGGAAGGCGAGA--UGAAAG--- ..((.((((((.(.((((..(((..(.((((.....)))).).....((((((.......))).)))...............)))...)))).).)))).))--.))...--- ( -31.60) >DroPer_CAF1 114557 102 + 1 GUUCCUCGGCGGUGCACUGAUCCGAGAGGCUGGUGAAACCGCAAAUGUCCGCAAAGAGGAUGCUGCAAGG-----GAAACGGGGAGGAG----CUUGAGAGACAGGCAGGA-- .((((((((((((.((((...((....))..))))..)))))....((((.......)))).........-----....)))))))..(----((((.....)))))....-- ( -35.20) >consensus GUUCCUCGGCGGUGCACUGAUCCGAGAGGCUGGUGAAACCGCAAAUGUCCGCAAAAAGGAUGCUGGAAAA_____G_AA_GGGGAAAAGGGGAAUUGUGAGA__UGCAGG___ .((((((.(((((.((((...((....))..))))..)))))....((((.......))))...................))))))........................... (-23.71 = -24.10 + 0.39)

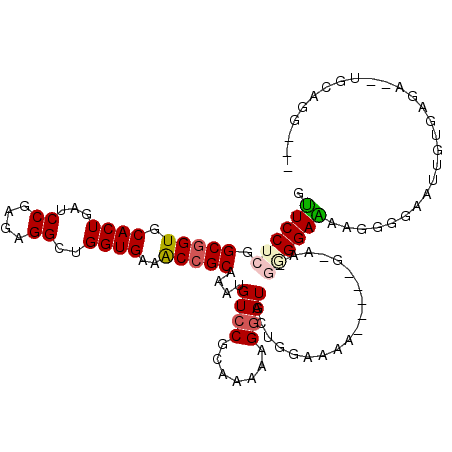
| Location | 16,479,646 – 16,479,752 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16479646 106 - 23771897 GGUCCUGCA--GCUUACAAUUCCCCUAUUCCUCUUCCC-----UUUUCCAGCAUCCUUUUUGCGGACAUCUGCGGUUUCACCAGCCUCUCGGAUCAGUGCACCGCCGAGGAAU .........--...............(((((((.....-----...(((.(((.......)))))).....(((((..(((...((....))....))).))))).))))))) ( -24.90) >DroPse_CAF1 111386 102 - 1 --UCCUGCCUGUCUCUCAAG----CUCCUCCCCGUUUC-----CCUUGCAGCAUCCUCUUUGCGGACAUUUGCGGUUUCACCAGCCUCUCGGAUCAGUGCACCGCCGAGGAAC --....((.((.....)).)----)(((((.((((...-----....)).(((.......)))))......(((((..(((...((....))....))).))))).))))).. ( -23.60) >DroEre_CAF1 93551 91 - 1 --------A--GCUCACAAUUCCCCUUUUCC------------AUUUCCAGCAUCCUUUUUGCGGACAUCUGCGGUUUCACCAGCCUCUCGGAUCAGUGCACCGCCGAGGAAC --------.--................((((------------...(((.(((.......))))))..((.(((((..(((...((....))....))).))))).)))))). ( -22.00) >DroYak_CAF1 96944 91 - 1 --------A--ACUCACAAUUCCUUUUUUCC------------UCUUCCAGCAUCCUUUUUGCGGACAUCUGCGGUUUCACCAGCCUCUCGGAUCAGUGCACCGCCGAGGAAC --------.--................((((------------((.(((.(((.......)))))).....(((((..(((...((....))....))).))))).)))))). ( -24.30) >DroAna_CAF1 102835 108 - 1 ---CUUUCA--UCUCGCCUUCCACCUUCUCCCCUUUCUGUGUGUUUUCCAGCAUUCUGUUUGCGGACAUUUGCGGCUUCACCAGCCUCUCGGAUCAGUGCACGGCGGAGGAGU ---......--.............(((((((.....(((((..((.(((.(((.((((....))))....)))((((.....))))....)))..))..))))).))))))). ( -32.10) >DroPer_CAF1 114557 102 - 1 --UCCUGCCUGUCUCUCAAG----CUCCUCCCCGUUUC-----CCUUGCAGCAUCCUCUUUGCGGACAUUUGCGGUUUCACCAGCCUCUCGGAUCAGUGCACCGCCGAGGAAC --....((.((.....)).)----)(((((.((((...-----....)).(((.......)))))......(((((..(((...((....))....))).))))).))))).. ( -23.60) >consensus ___CCUGCA__UCUCACAAUUCCCCUUCUCCCC_UU_C_____UCUUCCAGCAUCCUUUUUGCGGACAUCUGCGGUUUCACCAGCCUCUCGGAUCAGUGCACCGCCGAGGAAC .........................(((((................(((.(((.......)))))).....(((((..(((...((....))....))).))))).))))).. (-15.78 = -16.20 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:44 2006