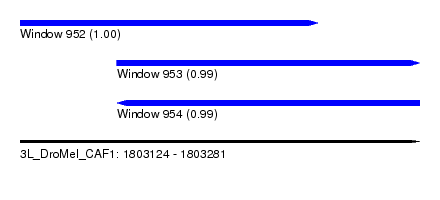
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,803,124 – 1,803,281 |
| Length | 157 |
| Max. P | 0.997358 |

| Location | 1,803,124 – 1,803,241 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.45 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -33.94 |
| Energy contribution | -34.70 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1803124 117 + 23771897 GACAGCGUCGAUCGGCGGACAGGUAUUGCUGCAACCGG---CGGCUCAUGCCAAUAUGCAUUGUGCAUAGCAAAUAAGUGCUUGUCAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGG .....((((....))))(((((((((.(((((.....)---))))..)))))....(((((((.(((.((((......))))))))))))))....))))..........((.....)). ( -36.90) >DroSec_CAF1 18486 117 + 1 GACAGCGCCGAUCGGCGGACAGGUAUUGCUGCAACCGG---CGGCUCAUGCCAAUAUGCAUUGUGCAUAGCAAAUAAGUGCUUGUCAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGG .....((((....))))(((((((((.(((((.....)---))))..)))))....(((((((.(((.((((......))))))))))))))....))))..........((.....)). ( -39.60) >DroSim_CAF1 18543 117 + 1 GACAGCGCCGAUCGGCGGACAGGUAUUGCUGCAACCGG---CGGCUCACGCCAAUAUGCAUUGUGCAUAGCAAAUAAGUGCUUGUCAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGG (((((((((....))))((((((((((..(((....((---((.....))))..(((((.....))))))))....)))))))))).........)))))..........((.....)). ( -41.10) >DroEre_CAF1 18720 114 + 1 GACAGCGUCGAUCGGCGGACAGGUAUUGCAGCAACCGG---CGGCUCAUGCCAAUAUGCAUUGUGCAUAGCAAAUAAGAGCU---CAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGG (((((((((....)))).....((((((.(((....((---((.....))))..(((((.....)))))..........)))---))))))....)))))..........((.....)). ( -30.50) >DroYak_CAF1 18930 120 + 1 GACAGCGCCGAUCGGCGGACAGGUAUUGCUGCAACCGGCGGCGGCUCAUGCCAAUAUGCAUUGUGCAUAGCAAAUAAGUGCUUGUCAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGG (((((((((....))))(((((((((((((((.....))))).(((.((((((((....)))).))))))).....)))))))))).........)))))..........((.....)). ( -42.30) >consensus GACAGCGCCGAUCGGCGGACAGGUAUUGCUGCAACCGG___CGGCUCAUGCCAAUAUGCAUUGUGCAUAGCAAAUAAGUGCUUGUCAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGG (((((((((....))))((((((((((..(((..........(((....)))..(((((.....))))))))....)))))))))).........)))))..........((.....)). (-33.94 = -34.70 + 0.76)

| Location | 1,803,162 – 1,803,281 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -34.66 |
| Consensus MFE | -29.72 |
| Energy contribution | -30.92 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1803162 119 + 23771897 -CGGCUCAUGCCAAUAUGCAUUGUGCAUAGCAAAUAAGUGCUUGUCAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGGGGAUAGUCGAGCCCGCCGACAGCUUAACCGCUGCCUCUUC -.((((..........(((((((.(((.((((......)))))))))))))).(((((.....)))))))))....((((((.........))).))).((((......))))....... ( -34.40) >DroSec_CAF1 18524 119 + 1 -CGGCUCAUGCCAAUAUGCAUUGUGCAUAGCAAAUAAGUGCUUGUCAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGGGGAUAGUCGAGCCCGCCGACAGCUUAACCGCUGCCUCUUC -.((((..........(((((((.(((.((((......)))))))))))))).(((((.....)))))))))....((((((.........))).))).((((......))))....... ( -34.40) >DroSim_CAF1 18581 119 + 1 -CGGCUCACGCCAAUAUGCAUUGUGCAUAGCAAAUAAGUGCUUGUCAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGGGGAUAUUCUAUCCCCCCGACAGCUUAACCGCUGCCUCUUC -.((((..........(((((((.(((.((((......)))))))))))))).(((((.....))))))))).....((((((((...)))))))).(.((((......)))).)..... ( -39.20) >DroEre_CAF1 18758 116 + 1 -CGGCUCAUGCCAAUAUGCAUUGUGCAUAGCAAAUAAGAGCU---CAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGGGCAUAGCCGAGCCCGCCGACAGCUUAACCGCCGCCUCUUC -.((((.(((((....(((((((.((.............)).---))))))).(((((.....)))))..((.....)))))))))))(((.(.((.(.........).)).).)))... ( -30.62) >DroYak_CAF1 18970 120 + 1 GCGGCUCAUGCCAAUAUGCAUUGUGCAUAGCAAAUAAGUGCUUGUCAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGGGGAUAGCCGAGCCCGCCGACAGCUUAACCGCUGCCUCUUC (((((((...((....(((((((.(((.((((......)))))))))))))).(((((.....))))).........))((.....))))).)))).(.((((......)))).)..... ( -34.70) >consensus _CGGCUCAUGCCAAUAUGCAUUGUGCAUAGCAAAUAAGUGCUUGUCAAUGCAAAAUUGUCAAUCAAUUAGCCAAAACGGGGGAUAGUCGAGCCCGCCGACAGCUUAACCGCUGCCUCUUC ..((((..........(((((((.(((.((((......)))))))))))))).(((((.....)))))))))....((((((.........))).))).((((......))))....... (-29.72 = -30.92 + 1.20)

| Location | 1,803,162 – 1,803,281 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -35.20 |
| Energy contribution | -35.08 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1803162 119 - 23771897 GAAGAGGCAGCGGUUAAGCUGUCGGCGGGCUCGACUAUCCCCCGUUUUGGCUAAUUGAUUGACAAUUUUGCAUUGACAAGCACUUAUUUGCUAUGCACAAUGCAUAUUGGCAUGAGCCG- .....((((((......))))))((((((...........))))))..((((...((.....((((..(((((((.((((((......)))).))..))))))).)))).))..)))).- ( -37.80) >DroSec_CAF1 18524 119 - 1 GAAGAGGCAGCGGUUAAGCUGUCGGCGGGCUCGACUAUCCCCCGUUUUGGCUAAUUGAUUGACAAUUUUGCAUUGACAAGCACUUAUUUGCUAUGCACAAUGCAUAUUGGCAUGAGCCG- .....((((((......))))))((((((...........))))))..((((...((.....((((..(((((((.((((((......)))).))..))))))).)))).))..)))).- ( -37.80) >DroSim_CAF1 18581 119 - 1 GAAGAGGCAGCGGUUAAGCUGUCGGGGGGAUAGAAUAUCCCCCGUUUUGGCUAAUUGAUUGACAAUUUUGCAUUGACAAGCACUUAUUUGCUAUGCACAAUGCAUAUUGGCGUGAGCCG- .....((((((......)))))).((((((((...)))))))).....((((...((.....((((..(((((((.((((((......)))).))..))))))).)))).))..)))).- ( -44.60) >DroEre_CAF1 18758 116 - 1 GAAGAGGCGGCGGUUAAGCUGUCGGCGGGCUCGGCUAUGCCCCGUUUUGGCUAAUUGAUUGACAAUUUUGCAUUG---AGCUCUUAUUUGCUAUGCACAAUGCAUAUUGGCAUGAGCCG- .....(.((((((.....)))))).).((((((((...(((.......)))...........((((..(((((((---.((.............)).))))))).)))))).)))))).- ( -39.02) >DroYak_CAF1 18970 120 - 1 GAAGAGGCAGCGGUUAAGCUGUCGGCGGGCUCGGCUAUCCCCCGUUUUGGCUAAUUGAUUGACAAUUUUGCAUUGACAAGCACUUAUUUGCUAUGCACAAUGCAUAUUGGCAUGAGCCGC .....((((((......)))))).((((.((((((((..........))))).......((.((((..(((((((.((((((......)))).))..))))))).)))).)).))))))) ( -38.90) >consensus GAAGAGGCAGCGGUUAAGCUGUCGGCGGGCUCGACUAUCCCCCGUUUUGGCUAAUUGAUUGACAAUUUUGCAUUGACAAGCACUUAUUUGCUAUGCACAAUGCAUAUUGGCAUGAGCCG_ .....((((((......))))))((((((...........))))))..((((...((.....((((..(((((((...(((........))).....))))))).)))).))..)))).. (-35.20 = -35.08 + -0.12)

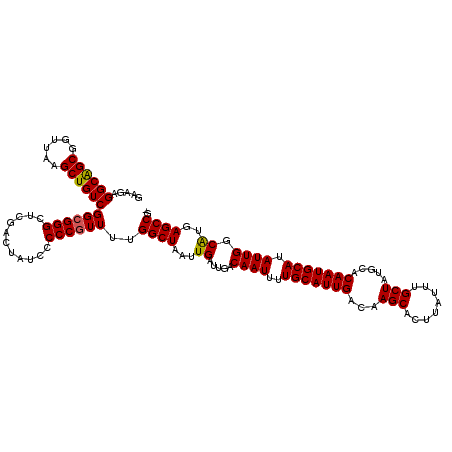
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:56 2006