| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,392,109 – 16,392,201 |
| Length | 92 |
| Max. P | 0.500000 |

| Location | 16,392,109 – 16,392,201 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -19.26 |
| Energy contribution | -18.85 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
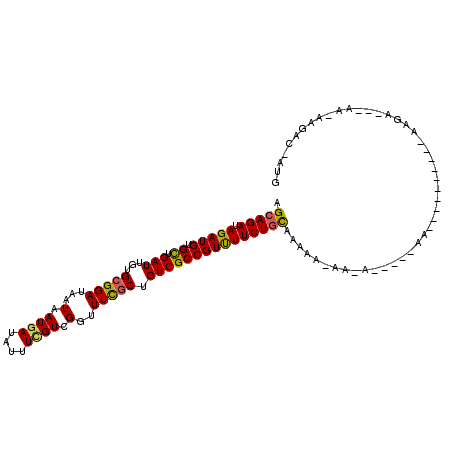
>3L_DroMel_CAF1 16392109 92 + 23771897 AGCAGAUAGAUGUGCUGAUUGUGCGGAUAAUAAUGAUAUUUCGUCGGUUUCGCUGUCGCCGUUUUCUACAAAAA---UG-----AA----------AAGA---AA-AAGUC-AUG .(.(((.(((((.((.(((...(((((...(.((((....)))).)..))))).))))))))))))).).....---..-----..----------....---..-.....-... ( -16.10) >DroVir_CAF1 7071 107 + 1 AGCAGAUAGAUGUGUUGAUUGUGCGGAUAAUAAUGAUAUUUGGUCGGUUUCGCUGUCGCCGUCUUCUGCAAAUAGAAUAGUUAGAAACG-------AGCCGUUGA-GUUGUGCUG .(((((.(((((.((.(((...(((((......((((.....))))..))))).)))))))))))))))....................-------(((((....-..)).))). ( -25.30) >DroGri_CAF1 7143 103 + 1 AGCAGAUAGAUGUGUUGAUUGUGCGGAUAAUAAUGAUAUUUUGUUGGUUUCGCUGUCGCCGUCUUCUGCAAAUAGA---GUUAGAAAUUAAAAACA-------CA-UAUAU-AUG .(((((.(((((.((.(((...(((((...(((..(....)..)))..))))).))))))))))))))).......---.................-------..-.....-... ( -24.30) >DroSec_CAF1 7134 94 + 1 AGCAGAUAGAUGUGCUGAUUGUGCGGAUAAUAAUGAUAUUUCGUCGGUUUUGCUGUCGCCGUUUUCUGUAAAAA-AAGA-----AG----------AAGA---AA-AAGCC-AUG .(((((.(((((.((.(((...(((((...(.((((....)))).)..))))).))))))))))))))).....-....-----..----------....---..-.....-... ( -18.40) >DroSim_CAF1 7198 95 + 1 AGCAGAUAGAUGUGCUGAUUGUGCGGAUAAUAAUGAUAUUUCGUCGGUUUCGCUGUCGCCGUUUUCUGUAAAAAAAAAA-----AG----------AAGA---AA-AAGCC-AUG .(((((.(((((.((.(((...(((((...(.((((....)))).)..))))).)))))))))))))))..........-----..----------....---..-.....-... ( -20.40) >DroAna_CAF1 7447 95 + 1 AGCAGAUAGAUGUGCUGAUUGUGGGGAUAAUAAUGAUAUUUCGUUGGUUUCGCUGUCGCCGUUUUCUGCAAAAACAA-G-----UC----------AAGA---AUUAUAAC-AUG .(((((.(((((.((.(((.(((..(....((((((....)))))).)..))).)))))))))))))))........-.-----..----------....---........-... ( -25.40) >consensus AGCAGAUAGAUGUGCUGAUUGUGCGGAUAAUAAUGAUAUUUCGUCGGUUUCGCUGUCGCCGUUUUCUGCAAAAA_AA_A_____AA__________AAGA___AA_AAGAC_AUG .(((((.(((((.((.(((...(((((...(.((((....)))).)..))))).))))))))))))))).............................................. (-19.26 = -18.85 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:20 2006