
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,384,569 – 16,384,664 |
| Length | 95 |
| Max. P | 0.990876 |

| Location | 16,384,569 – 16,384,664 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -22.28 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
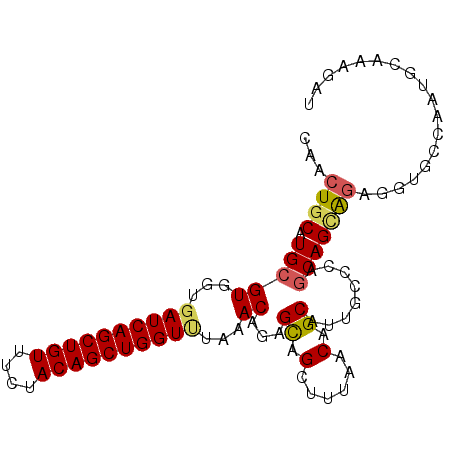
>3L_DroMel_CAF1 16384569 95 + 23771897 CAAAUGCAUGCGUGGUGAUCAGCUGUUUCUACAGCUGGUUUAAACAGAGCAGAUUGAACAGCAUUGCCAAACAGCAGAGGUGCCAAUGCAAAGAU ....(((.(((.((..((((((((((....))))))))))....))..))).((((..((.(.((((......)))).).)).)))))))..... ( -27.40) >DroSec_CAF1 1822 95 + 1 CAACUGCAUGCGUGGUGAUCAGCUGUUUAUACAGCUGGUUUAAACAGAGUAGCUUUAACAGCAUUGUCCAGCAGCAGAGGUGCCAAUGUAAAGAU ...((((.(((((...((((((((((....))))))))))...))......(((.....)))........))))))).................. ( -28.20) >DroSim_CAF1 1823 95 + 1 CAACUGCAUGCGUGGUGAUCAGCUGUUUCUACAGCUGGUUUAAACAGAGCAGCUUUAACAGCAUUGCUCAGCAGUAGAGGUGCCAAUGCAAAAAU ...((((.(((((...((((((((((....))))))))))...)).((((((((.....)))..))))).)))))))...(((....)))..... ( -34.10) >DroEre_CAF1 1822 89 + 1 CAACUGCAUGCGUGGUGAUCAGCUGUUUCUACAGCUGGUCUUAACAGGGCAGUCCUAACAGCAUUACUUAGCAGCAGUG------AUGCCAAUAU ..(((((....((...((((((((((....))))))))))...))...))))).......((((((((.......))))------))))...... ( -32.30) >DroYak_CAF1 1821 92 + 1 CAACUGCAUGCGUGGUGAUCAGCUGUUUCUACAGCUGGUACCAACAGAGCAGUCCUAACAGCAUUACCUAGCAGAGGUG---UAAGUGCAAAGUU ..(((((.((..((((.(((((((((....))))))))))))).))..))))).......(((((((((.....)))))---...))))...... ( -33.00) >consensus CAACUGCAUGCGUGGUGAUCAGCUGUUUCUACAGCUGGUUUAAACAGAGCAGCUUUAACAGCAUUGCCCAGCAGCAGAGGUGCCAAUGCAAAGAU ...((((.(((((...((((((((((....))))))))))...))...((.(......).))........))))))).................. (-22.28 = -22.52 + 0.24)

| Location | 16,384,569 – 16,384,664 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.86 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16384569 95 - 23771897 AUCUUUGCAUUGGCACCUCUGCUGUUUGGCAAUGCUGUUCAAUCUGCUCUGUUUAAACCAGCUGUAGAAACAGCUGAUCACCACGCAUGCAUUUG .....(((((.((((....))))((.(((....((((((...(((((.(((.......)))..)))))))))))......))).))))))).... ( -24.30) >DroSec_CAF1 1822 95 - 1 AUCUUUACAUUGGCACCUCUGCUGCUGGACAAUGCUGUUAAAGCUACUCUGUUUAAACCAGCUGUAUAAACAGCUGAUCACCACGCAUGCAGUUG ..................((((((((((........(((.((((......)))).)))(((((((....)))))))....))).))).))))... ( -25.90) >DroSim_CAF1 1823 95 - 1 AUUUUUGCAUUGGCACCUCUACUGCUGAGCAAUGCUGUUAAAGCUGCUCUGUUUAAACCAGCUGUAGAAACAGCUGAUCACCACGCAUGCAGUUG ....(((((((((..........((.(((((..(((.....)))))))).))......(((((((....)))))))....)))...))))))... ( -27.30) >DroEre_CAF1 1822 89 - 1 AUAUUGGCAU------CACUGCUGCUAAGUAAUGCUGUUAGGACUGCCCUGUUAAGACCAGCUGUAGAAACAGCUGAUCACCACGCAUGCAGUUG ...((((((.------((.((((....)))).)).))))))((((((..(((...((.(((((((....))))))).))...)))...)))))). ( -27.80) >DroYak_CAF1 1821 92 - 1 AACUUUGCACUUA---CACCUCUGCUAGGUAAUGCUGUUAGGACUGCUCUGUUGGUACCAGCUGUAGAAACAGCUGAUCACCACGCAUGCAGUUG (((...(((....---.((((.....))))..))).)))..((((((..(((((((..(((((((....)))))))...)))).))).)))))). ( -28.50) >consensus AUCUUUGCAUUGGCACCUCUGCUGCUAGGCAAUGCUGUUAAAACUGCUCUGUUUAAACCAGCUGUAGAAACAGCUGAUCACCACGCAUGCAGUUG ....................(((((......((((.(((.((((......)))).)))(((((((....)))))))........))))))))).. (-18.46 = -18.86 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:13 2006