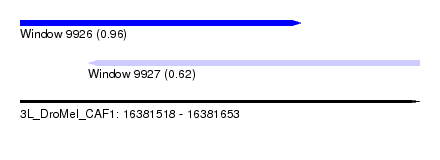
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,381,518 – 16,381,653 |
| Length | 135 |
| Max. P | 0.956508 |

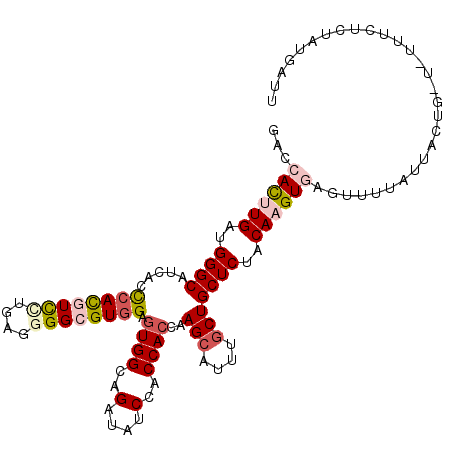
| Location | 16,381,518 – 16,381,613 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 72.91 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -13.74 |
| Energy contribution | -13.77 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16381518 95 + 23771897 CAUAUAUAUCUCUA------AAUGGACAGAAUCAUAGAGAAAAAACAUCAAUGAAACCCACUUGUAGAGCAGCAAAUGCUUGGUGGUGGAUAUCUGCCACU ..............------..(((.((((.((((.((.........)).))))...(((((....(((((.....)))))...)))))...))))))).. ( -23.30) >DroSec_CAF1 11181 84 + 1 CACA----------------UGUGGAAAGAAUCAUAGAGAAA-UACAACAAUGAAACCCACUUGUAGAGCAGCAAAUGCUUGGUGGUGGAUAUCUGCCACU .(((----------------.((((......((((.......-.......))))...)))).))).(((((.....)))))(((((..(....)..))))) ( -21.14) >DroSim_CAF1 11164 84 + 1 CAUA----------------AGUGGACAGAAUCAUAGAGAAA-AACAACAAUGAAACCCACUUGUAGAGCAGCAAAUGCUUGGUGGUGGAUAUCUGCCACU ....----------------(((((.((((.((((.......-.......))))...(((((....(((((.....)))))...)))))...))))))))) ( -24.74) >DroEre_CAF1 11246 90 + 1 CACAU----AUUUG------AGUACACAAUAUCAUAGAGCAA-AGCAGUAAUAAAACGCACUUGUAGAGCAGCAAAUGCUUGGUGGUGGAUAUCUGCCAUU .....----.....------................(((((.-.((.((......))))..((((......)))).)))))(((((..(....)..))))) ( -15.50) >DroYak_CAF1 35927 87 + 1 CAUAU----CUCUA------AGUACAAUUUAUUA---AGGAG-AGCAGUAUAAAAACGAACUUGUAGAGCAGCAAAUGCUUGGUGGUGGAUAUCUGCCACU ....(----((((.------((((.....)))).---.))))-)((..(((((........)))))..))(((....))).(((((..(....)..))))) ( -19.00) >DroPer_CAF1 17689 90 + 1 CAAAU----AUUAUCGUAGAAAUGGAUUGAGACUGAGAC-------AGGGGAAAAUCUCACUUGUAGAGCAGCAAAUGCUUUGUGGUCGAUAUCUGCCACU .....----......(((((.((.((((.........((-------(((.((.....)).)))))((((((.....))))))..)))).)).))))).... ( -19.50) >consensus CAUAU_____U_U_______AGUGGACAGAAUCAUAGAGAAA_AACAGCAAUAAAACCCACUUGUAGAGCAGCAAAUGCUUGGUGGUGGAUAUCUGCCACU ..................................................................(((((.....))))).(((((((....))))))). (-13.74 = -13.77 + 0.03)

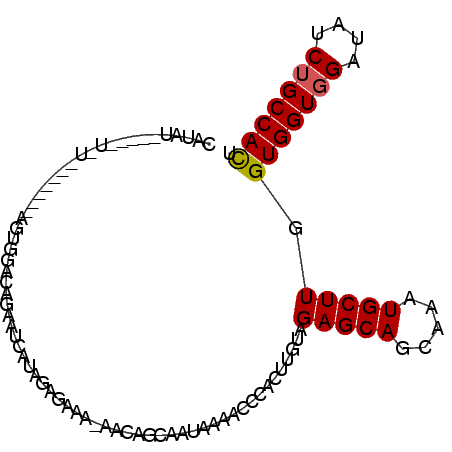
| Location | 16,381,541 – 16,381,653 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -22.84 |
| Energy contribution | -23.82 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16381541 112 - 23771897 GACCAUUUGAUGGGCAUCACCCACGUCCUGAGGGGCGUGGAGUGGCAGAUAUCCACCACCAAGCAUUUGCUGCUCUACAAGUGGGUUUCAUUGAUGUUUUUUCUCUAUGAUU ...(((..((.(((((((((((((((((....))))((((((..((((((..............))))))..))))))..))))))......)))))))..))...)))... ( -40.94) >DroSec_CAF1 11194 111 - 1 GACCACUUGAUGGGCAUUACCCAUGUCCUGAGGGGCGUGGAGUGGCAGAUAUCCACCACCAAGCAUUUGCUGCUCUACAAGUGGGUUUCAUUGUUGUA-UUUCUCUAUGAUU ..((((((.(((((.....)))))((((....))))((((((..((((((..............))))))..))))))))))))..............-............. ( -37.14) >DroEre_CAF1 11265 111 - 1 GACCAUUUGAUGGGCAUUACCCACGUCUUGAGGGGCGUGGAAUGGCAGAUAUCCACCACCAAGCAUUUGCUGCUCUACAAGUGCGUUUUAUUACUGCU-UUGCUCUAUGAUA ...(((.....(((((....((((((((....))))))))...(((((.((...........(((((((........))))))).......)))))))-.))))).)))... ( -29.77) >DroYak_CAF1 35946 108 - 1 GACCACUUGAUGGGCAUUACCCACGUCUUGAGGGGCGUGGAGUGGCAGAUAUCCACCACCAAGCAUUUGCUGCUCUACAAGUUCGUUUUUAUACUGCU-CUCCU---UAAUA ..........((((.....))))....((((((((.((((((..((((((..............))))))..)))))).(((..((......)).)))-)))))---))).. ( -30.54) >DroAna_CAF1 33726 109 - 1 GACCAUCUGAUGGGCAUCACUCACGUCCUGAGAGGAGUGGAGUGGCAGAUAUCCACCACCAAGCACCUCCUGCUCUACAAGUAAGUAUUUCAAAAA---CCUCCCUAAACUG ..((((...)))).....(((.((.....(((((((((((.((((.......))))..))).....))))).))).....)).)))..........---............. ( -20.70) >DroPer_CAF1 17714 105 - 1 GACCACCUGAUGGGCAUCACCCAUGUUCUGCGAGGAGUUGAGUGGCAGAUAUCGACCACAAAGCAUUUGCUGCUCUACAAGUGAGAUUUUCCCCU-------GUCUCAGUCU (((..(((.(((((.....)))))(.....).))).((.(((..((((((..............))))))..))).))...((((((........-------))))))))). ( -28.94) >consensus GACCACUUGAUGGGCAUCACCCACGUCCUGAGGGGCGUGGAGUGGCAGAUAUCCACCACCAAGCAUUUGCUGCUCUACAAGUGAGUUUUAUUACUG_U_UUUCUCUAUGAUU ...((((((..((((.....((((((((....)))))))).((((..(....)..))))..(((....)))))))..))))))............................. (-22.84 = -23.82 + 0.97)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:10 2006