
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,368,607 – 16,368,737 |
| Length | 130 |
| Max. P | 0.991791 |

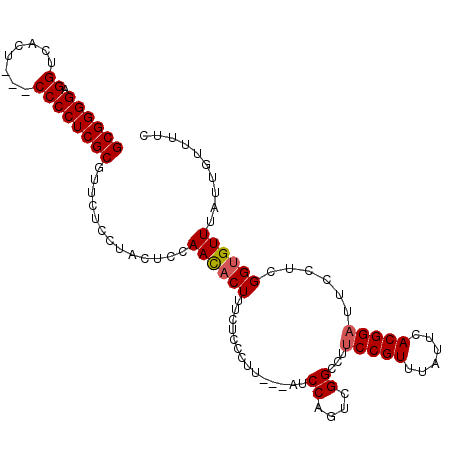
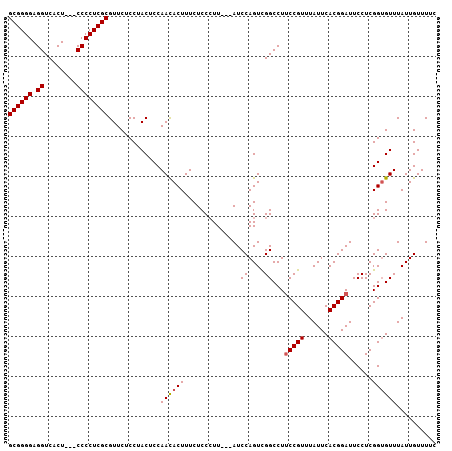
| Location | 16,368,607 – 16,368,714 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 94.71 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -19.49 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16368607 107 - 23771897 GCGGGGAGGUCACUACCCCCCUCGCGUUCUCCUACUCCAACACUUUCUCCCUUUACAUCCAGUUGGCCUUCCGUUUAUUCACGGAUUCCUCGGUGUUUAUUGUUUUC ((((((.(((....))).))).))).............((((((.....((...((.....)).))...(((((......)))))......)))))).......... ( -23.30) >DroSec_CAF1 8727 101 - 1 GCGGGGAGGUCACU---CCCCUCGCGUUCUCCUACUCCAACACUUUCUCCCUU---AUCCAGUCGGCCUUCCGUUUAUUCACGGAUUCCUCGGAGUUUAUUGUUUUC ((((((((....))---))))..))........(((((...(((.........---....))).((...(((((......)))))..))..)))))........... ( -25.72) >DroSim_CAF1 8857 101 - 1 GCGGGGAGGUCACU---CCCCUCGCGUUCUCCUACUCCAACACUUUCUCCCUU---AUCCAGUCGGCCUUCCGUUUAUUCACGGAUUCCUCGGUGUUUAUUGUUUUC ((((((((....))---))))..)).............((((((......((.---....))..((...(((((......)))))..))..)))))).......... ( -23.20) >DroEre_CAF1 8619 100 - 1 GCGGGGAGGUCACU---CCCCUCGCGUUCUCCUACUCCAAUACUUUCUCC-UU---AUCCAGUCGGCCUCCCGUUUAUUCACGGAUUCCUCGGUGUUUAUUGUUUUC ((((((((....))---))))..))............(((((....(.((-..---..((....))....((((......)))).......)).)..)))))..... ( -21.00) >consensus GCGGGGAGGUCACU___CCCCUCGCGUUCUCCUACUCCAACACUUUCUCCCUU___AUCCAGUCGGCCUUCCGUUUAUUCACGGAUUCCUCGGUGUUUAUUGUUUUC ((((((.((........)))))))).............((((((..............((....))...(((((......)))))......)))))).......... (-19.49 = -19.80 + 0.31)

| Location | 16,368,640 – 16,368,737 |
|---|---|
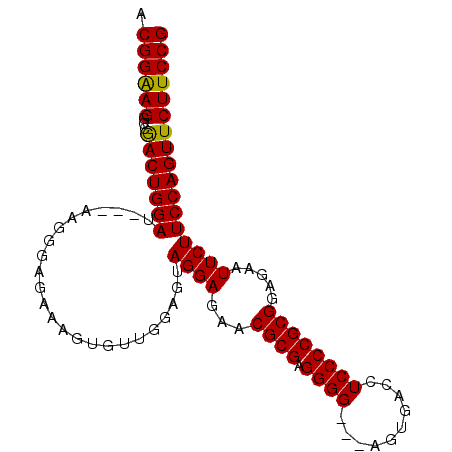
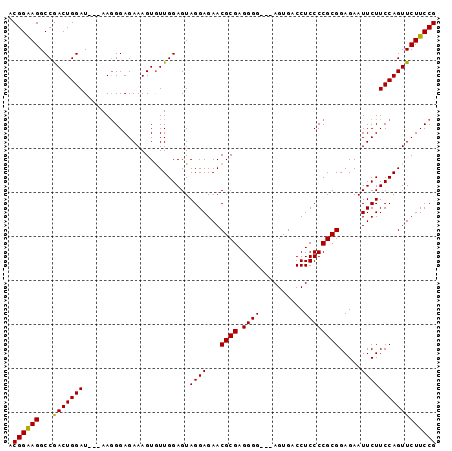
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.68 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -27.27 |
| Energy contribution | -26.90 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16368640 97 + 23771897 ACGGAAGGCCAACUGGAUGUAAAGGGAGAAAGUGUUGGAGUAGGAGAACGCGAGGGGGGUAGUGACCUCCCCGCGGAGAAUUCUUCCAGUUCUUCCG .(....).((....)).......(((((((....(((((..((((...((((.(((((((....))))))))))).....)))))))))))))))). ( -37.10) >DroSec_CAF1 8760 91 + 1 ACGGAAGGCCGACUGGAU---AAGGGAGAAAGUGUUGGAGUAGGAGAACGCGAGGGG---AGUGACCUCCCCGCGGAGAAUUCUUCCAGUUCUUCCG .(((((((((....))..---..(((((((.(((((..........)))))(.((((---((....)))))).)......)))))))...))))))) ( -33.20) >DroSim_CAF1 8890 91 + 1 ACGGAAGGCCGACUGGAU---AAGGGAGAAAGUGUUGGAGUAGGAGAACGCGAGGGG---AGUGACCUCCCCGCGGAGAAUUCUUCCAGUUCUUCCG .(((((((((....))..---..(((((((.(((((..........)))))(.((((---((....)))))).)......)))))))...))))))) ( -33.20) >DroEre_CAF1 8652 90 + 1 ACGGGAGGCCGACUGGAU---AA-GGAGAAAGUAUUGGAGUAGGAGAACGCGAGGGG---AGUGACCUCCCCGCGGAGAAUUCUUCCAGUUCUUCCG .(((((((.(..((....---.)-)..).....((((((..((((...(((..((((---((....))))))))).....))))))))))))))))) ( -30.90) >consensus ACGGAAGGCCGACUGGAU___AAGGGAGAAAGUGUUGGAGUAGGAGAACGCGAGGGG___AGUGACCUCCCCGCGGAGAAUUCUUCCAGUUCUUCCG .((((((...(((((((........................((((...((((.((((..........)))))))).....))))))))))))))))) (-27.27 = -26.90 + -0.37)

| Location | 16,368,640 – 16,368,737 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 94.68 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -27.20 |
| Energy contribution | -27.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16368640 97 - 23771897 CGGAAGAACUGGAAGAAUUCUCCGCGGGGAGGUCACUACCCCCCUCGCGUUCUCCUACUCCAACACUUUCUCCCUUUACAUCCAGUUGGCCUUCCGU (((((((((((((((....)).(((((((.(((....))).))).))))...............................)))))))...)))))). ( -30.30) >DroSec_CAF1 8760 91 - 1 CGGAAGAACUGGAAGAAUUCUCCGCGGGGAGGUCACU---CCCCUCGCGUUCUCCUACUCCAACACUUUCUCCCUU---AUCCAGUCGGCCUUCCGU ((((((.((((((((....)).(((((((((....))---))))..)))...........................---.))))))....)))))). ( -29.40) >DroSim_CAF1 8890 91 - 1 CGGAAGAACUGGAAGAAUUCUCCGCGGGGAGGUCACU---CCCCUCGCGUUCUCCUACUCCAACACUUUCUCCCUU---AUCCAGUCGGCCUUCCGU ((((((.((((((((....)).(((((((((....))---))))..)))...........................---.))))))....)))))). ( -29.40) >DroEre_CAF1 8652 90 - 1 CGGAAGAACUGGAAGAAUUCUCCGCGGGGAGGUCACU---CCCCUCGCGUUCUCCUACUCCAAUACUUUCUCC-UU---AUCCAGUCGGCCUCCCGU .((..((.((((((((((....((.((((((....))---)))).)).)))))....................-..---.)))))))..))...... ( -25.60) >consensus CGGAAGAACUGGAAGAAUUCUCCGCGGGGAGGUCACU___CCCCUCGCGUUCUCCUACUCCAACACUUUCUCCCUU___AUCCAGUCGGCCUUCCGU ((((((.((((((((....)).(((((((.((........)))))))))...............................))))))....)))))). (-27.20 = -27.45 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:07 2006