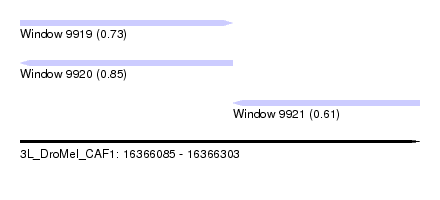
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,366,085 – 16,366,303 |
| Length | 218 |
| Max. P | 0.849042 |

| Location | 16,366,085 – 16,366,201 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.53 |
| Mean single sequence MFE | -38.44 |
| Consensus MFE | -34.12 |
| Energy contribution | -34.49 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16366085 116 + 23771897 UGUGUUCCCGGGUAAACGGGACAUUCCCCGGAUAAGCGGAUUGCUCCGAGUAACGGUGAAAUGAGCAGGAGCGAAUCCGAACCGAGCAGUUGCAGUGAACAGCGGCAACUGGAUGA .((((.((((......))))))))....(((.....(((((((((((..((.............)).))))).))))))..)))..(((((((.((.....)).)))))))..... ( -41.02) >DroSec_CAF1 6248 116 + 1 UGUGUUCCUGGAUAAACGGGACUUUCCCCGGAUAAGCGGAUUGCUCCGAGUAACGGUGAAAAGAGCAGGAGCGAAUCCGAACCGAGCAGUUGCAGUGAACAGCGGCAACUGGAUGA .....(((((......))))).......(((.....(((((((((((..((.............)).))))).))))))..)))..(((((((.((.....)).)))))))..... ( -37.32) >DroSim_CAF1 6362 116 + 1 UGUGUUCCCGGAUAAACGGGACUUUCCCCGGAUAAGCGGAUUGCUCCGAGUAACGGUGAAAAGAGCAGGAGCGAAUCCGAACCGAGCAGUUGCAGUGAACAGCGGCAACUGGAUGA .....(((((......))))).......(((.....(((((((((((..((.............)).))))).))))))..)))..(((((((.((.....)).)))))))..... ( -39.52) >DroEre_CAF1 6226 115 + 1 GGUUUU-CCGGAUAAACGGGAUACUCCCCGGAUAAACGGACUGCGUCAAGUAACGGUAAAAAGAGCAGGAGCGAAUGCGAACCGAGCAGUUGCAGUGAACAGCGGCAACUGGAUUA ((((((-(((..((..((((......))))..))..))))..((((...((..(.((.......)).)..))..)))))))))...(((((((.((.....)).)))))))..... ( -35.90) >consensus UGUGUUCCCGGAUAAACGGGACAUUCCCCGGAUAAGCGGAUUGCUCCGAGUAACGGUGAAAAGAGCAGGAGCGAAUCCGAACCGAGCAGUUGCAGUGAACAGCGGCAACUGGAUGA ........(((.....((((......))))......(((((((((((..((.............)).))))).))))))..)))..(((((((.((.....)).)))))))..... (-34.12 = -34.49 + 0.38)


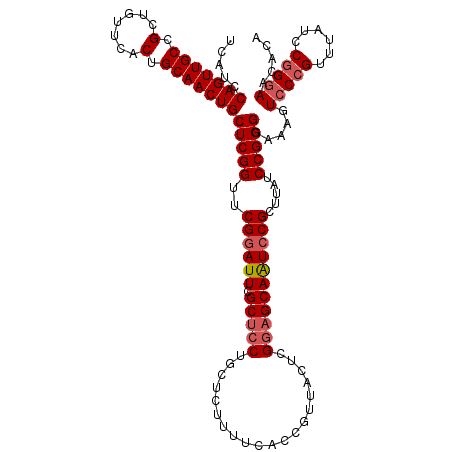
| Location | 16,366,085 – 16,366,201 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.53 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -30.41 |
| Energy contribution | -31.47 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16366085 116 - 23771897 UCAUCCAGUUGCCGCUGUUCACUGCAACUGCUCGGUUCGGAUUCGCUCCUGCUCAUUUCACCGUUACUCGGAGCAAUCCGCUUAUCCGGGGAAUGUCCCGUUUACCCGGGAACACA .....(((((((.(.......).)))))))(((((..((((((.(((((....................))))))))))).....)))))...(((((((......)))).))).. ( -37.65) >DroSec_CAF1 6248 116 - 1 UCAUCCAGUUGCCGCUGUUCACUGCAACUGCUCGGUUCGGAUUCGCUCCUGCUCUUUUCACCGUUACUCGGAGCAAUCCGCUUAUCCGGGGAAAGUCCCGUUUAUCCAGGAACACA .....(((((((.(.......).)))))))....(((((((((.(((((....................))))))))))(..((..(((((....)))))..))..)..))))... ( -33.85) >DroSim_CAF1 6362 116 - 1 UCAUCCAGUUGCCGCUGUUCACUGCAACUGCUCGGUUCGGAUUCGCUCCUGCUCUUUUCACCGUUACUCGGAGCAAUCCGCUUAUCCGGGGAAAGUCCCGUUUAUCCGGGAACACA .....(((((((.(.......).)))))))(((((..((((((.(((((....................))))))))))).....))))).....(((((......)))))..... ( -37.35) >DroEre_CAF1 6226 115 - 1 UAAUCCAGUUGCCGCUGUUCACUGCAACUGCUCGGUUCGCAUUCGCUCCUGCUCUUUUUACCGUUACUUGACGCAGUCCGUUUAUCCGGGGAGUAUCCCGUUUAUCCGG-AAAACC .....(((((((.(.......).)))))))...((((.((....))..((((((...............)).))))((((..((..((((......))))..))..)))-).)))) ( -30.06) >consensus UCAUCCAGUUGCCGCUGUUCACUGCAACUGCUCGGUUCGGAUUCGCUCCUGCUCUUUUCACCGUUACUCGGAGCAAUCCGCUUAUCCGGGGAAAGUCCCGUUUAUCCGGGAACACA .....(((((((.(.......).)))))))(((((..((((((.(((((....................))))))))))).....))))).....(((((......)))))..... (-30.41 = -31.47 + 1.06)


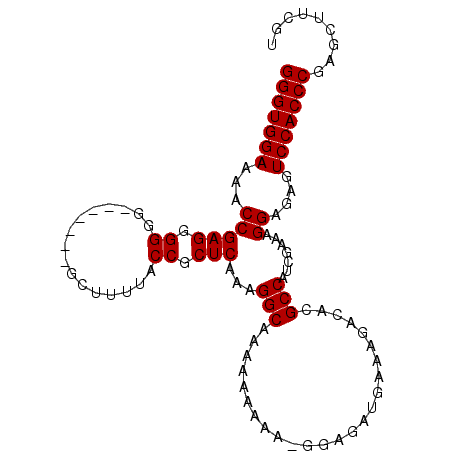
| Location | 16,366,201 – 16,366,303 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 92.69 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -26.10 |
| Energy contribution | -26.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16366201 102 - 23771897 GGGUGGAAAACCGAGGGGGGCAGAGGUGCUUUUACCGCUCAAAGGCAAAAAAAAG-GGAGAUGAAAGACACGCCAUCGAAAGGAGAGUCCACCCGAGCUCCGU (((((((...((..((..(((....(((((((((.(.(((..............)-)).).)))))).)))))).))....))....)))))))......... ( -29.24) >DroSec_CAF1 6364 95 - 1 GGGUGGAAAACCGAGGGGGG-------GCUUUUACCGCUCAAAGGCAAAAAAAAA-GGAGAUGAAAGACACGCCAUCGAAAGGAAAGUCCACCCGAGCUUCGU (((((((...(((((.((..-------.......)).)))...(((.........-...............))).......))....)))))))......... ( -26.76) >DroSim_CAF1 6478 96 - 1 GGGUGGAAAACCGAGGGGGG-------GCUUUUACCGCUCAAAGGCAAAAAAAAAGGGAGAUGAAAGACACGCCAUCGAAAGGAGAGUCCACCCGAGCUUCGU (((((((...(((((.((..-------.......)).)))...(((.........................))).......))....)))))))......... ( -26.71) >consensus GGGUGGAAAACCGAGGGGGG_______GCUUUUACCGCUCAAAGGCAAAAAAAAA_GGAGAUGAAAGACACGCCAUCGAAAGGAGAGUCCACCCGAGCUUCGU (((((((...(((((.((................)).)))...(((.........................))).......))....)))))))......... (-26.10 = -26.10 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:04 2006