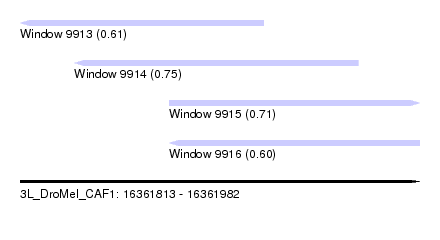
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,361,813 – 16,361,982 |
| Length | 169 |
| Max. P | 0.751426 |

| Location | 16,361,813 – 16,361,916 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.13 |
| Mean single sequence MFE | -20.45 |
| Consensus MFE | -15.93 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16361813 103 - 23771897 CAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCUCGAUCGGCUUAAUUAUCUUGUAUUUCGACAAGAUCAUUGCUAUCAGCUCGAAUUAGUGAUUC-----------------U .(((((((.(((.((((((....................))))))(((..(((.(((((((......))))))).))))))......)))...)))))))..-----------------. ( -20.85) >DroSec_CAF1 2074 103 - 1 CAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCUCGAUCGGCUUAAUUAUCUUGUAUUUCGACAAGAUCAUUGCUAUCAGCUCGAAUUGGUGAUUC-----------------U ....((((((((.((((((....................))))))((((((((.(((((((......))))))).)))).....))))....))))))))..-----------------. ( -23.15) >DroSim_CAF1 2078 103 - 1 CAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCUCGAUCGGCUUAAUUAUCUUGUAUUUCGACAAGAUCAUUGCUAUCAGCUCGAAUUAGUGAUUC-----------------U .(((((((.(((.((((((....................))))))(((..(((.(((((((......))))))).))))))......)))...)))))))..-----------------. ( -20.85) >DroEre_CAF1 2004 119 - 1 CAUCAUUAUGGCAGAUCGAACAUCAUGCAUUCAAUUAUCUUGAUCGGCUUCAUUAUCUUGUAUUUCGACAAGAUCAUUGCUAUCAUCUCGAACUAGUGAUUCAUAAUACUUCC-CAUCAU .(((((((..((((((.....))).)))...((((.((((((.((((...((......))....)))))))))).))))..............))))))).............-...... ( -19.20) >DroYak_CAF1 1927 120 - 1 CAUCAUUAUGGAAGAUCGAACAUCAUGCACUCAAUUAUCUUGAUCGGCUUCAUUAUCUGGUAUUUCGACAAGAUCAUUGCUAUCAUCUCGAAUGAGUUAUUCAUAAUACAUCCACAUCAU ........((((...((((.....(((....((((.((((((.((((..(((.....)))....)))))))))).))))....))).))))(((((...)))))......))))...... ( -18.20) >consensus CAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCUCGAUCGGCUUAAUUAUCUUGUAUUUCGACAAGAUCAUUGCUAUCAGCUCGAAUUAGUGAUUC_________________U .(((((((.....((((((....................))))))(((...((.(((((((......))))))).)).)))............))))))).................... (-15.93 = -16.13 + 0.20)

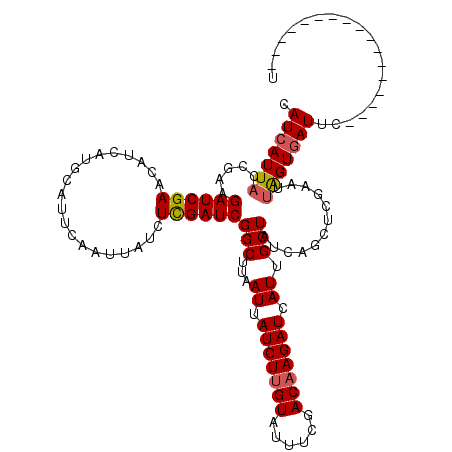
| Location | 16,361,836 – 16,361,956 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -30.81 |
| Consensus MFE | -23.77 |
| Energy contribution | -24.13 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16361836 120 - 23771897 UCUGCGGAUUGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCUCGAUCGGCUUAAUUAUCUUGUAUUUCGACAAGAUCAUUGC ....(((..((((...(((((((.(.....).)))))))..))))...)))..((((((....................))))))....((((.(((((((......))))))).)))). ( -29.55) >DroSec_CAF1 2097 120 - 1 UCUGCGGAUCGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCUCGAUCGGCUUAAUUAUCUUGUAUUUCGACAAGAUCAUUGC ...((.((((((....(((((((.(.....).)))))))...........((.(((.(((.........))).))).)))))))).)).((((.(((((((......))))))).)))). ( -29.20) >DroSim_CAF1 2101 120 - 1 UCCGCGGAUCGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCUCGAUCGGCUUAAUUAUCUUGUAUUUCGACAAGAUCAUUGC ...((.((((((....(((((((.(.....).)))))))...........((.(((.(((.........))).))).)))))))).)).((((.(((((((......))))))).)))). ( -29.50) >DroEre_CAF1 2043 120 - 1 UCUGCAGUUUGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUAUGGCAGAUCGAACAUCAUGCAUUCAAUUAUCUUGAUCGGCUUCAUUAUCUUGUAUUUCGACAAGAUCAUUGC ..((((((((((((...((((((.(.....).))))))(((((....))))).))))))))....))))..((((.((((((.((((...((......))....)))))))))).)))). ( -33.90) >DroYak_CAF1 1967 120 - 1 UCUGCAGUUUGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUAUGGAAGAUCGAACAUCAUGCACUCAAUUAUCUUGAUCGGCUUCAUUAUCUGGUAUUUCGACAAGAUCAUUGC ..((((((((((((..(((((((.(.....).)))))))((((....))))..))))))))....))))..((((.((((((.((((..(((.....)))....)))))))))).)))). ( -31.90) >consensus UCUGCGGAUUGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCUCGAUCGGCUUAAUUAUCUUGUAUUUCGACAAGAUCAUUGC ...((.....(((...(((((((.(.....).)))))))..))).........((((((....................)))))).)).((((.(((((((......))))))).)))). (-23.77 = -24.13 + 0.36)

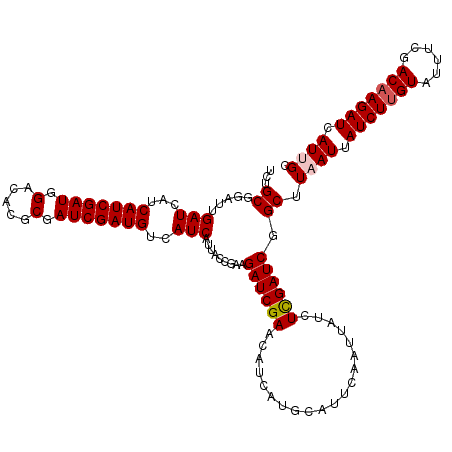
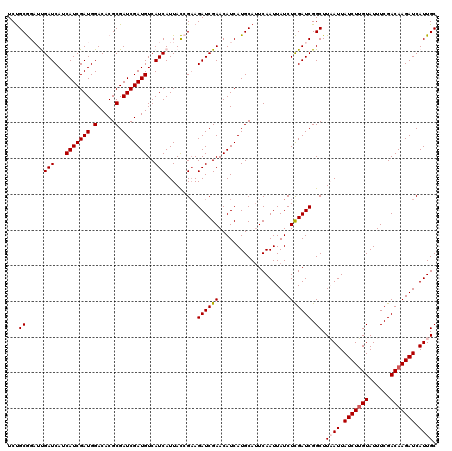
| Location | 16,361,876 – 16,361,982 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 96.04 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16361876 106 + 23771897 AGAUAAUUGAAUGCAUGAUGUUCGAUCUUCGGUAAUGAUGACAUCGAUCGCGUGUCCAUCGAUGAUGAUCAAUCCGCAGACAGUAUCGACAUGUUAAUUAUUAGCU .((((((((...(((((.((..(..(((.(((...((((.((((((((.(.....).))))))).).))))..))).)))..)...)).))))))))))))).... ( -26.80) >DroSec_CAF1 2137 106 + 1 AGAUAAUUGAAUGCAUGAUGUUCGAUCUUCGGUAAUGAUGACAUCGAUCGCGUGUCCAUCGAUGAUGAUCGAUCCGCAGACAGUAUCGACAUGUUAAUUAUUAGCU .((..(((((((.......)))))))..))(.((((((((((((((((.((.((((.((((((....)))))).....)))))))))))..)))).))))))).). ( -27.40) >DroSim_CAF1 2141 106 + 1 AGAUAAUUGAAUGCAUGAUGUUCGAUCUUCGGUAAUGAUGACAUCGAUCGCGUGUCCAUCGAUGAUGAUCGAUCCGCGGACAGUAUCGACAUGUUAAUUAUUAGCU .((..(((((((.......)))))))..))(.((((((((((((((((.((.(((((((((((....))))))....))))))))))))..)))).))))))).). ( -31.30) >DroEre_CAF1 2083 106 + 1 AGAUAAUUGAAUGCAUGAUGUUCGAUCUGCCAUAAUGAUGACAUCGAUCGCGUGUCCAUCGAUGAUGAUCAAACUGCAGACAGUAUCGACAUGUUAAUUAUUAGCU .((((((((...(((((.((..(..(((((.....((((.((((((((.(.....).))))))).).))))....)))))..)...)).))))))))))))).... ( -26.40) >DroYak_CAF1 2007 106 + 1 AGAUAAUUGAGUGCAUGAUGUUCGAUCUUCCAUAAUGAUGACAUCGAUCGCGUGUCCAUCGAUGAUGAUCAAACUGCAGACAGUAUCGACAUGUUAAUUAUUAGCU .((((((((...(((((....(((((.........((((.((((((((.(.....).))))))).).)))).((((....)))))))))))))))))))))).... ( -23.80) >consensus AGAUAAUUGAAUGCAUGAUGUUCGAUCUUCGGUAAUGAUGACAUCGAUCGCGUGUCCAUCGAUGAUGAUCAAUCCGCAGACAGUAUCGACAUGUUAAUUAUUAGCU .((((((((...(((((.((..(..(((.(((...((((.((((((((.(.....).))))))).).))))..))).)))..)...)).))))))))))))).... (-23.20 = -23.60 + 0.40)


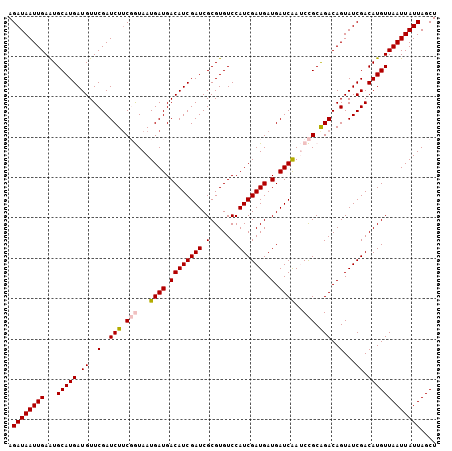
| Location | 16,361,876 – 16,361,982 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 96.04 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -19.82 |
| Energy contribution | -19.54 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16361876 106 - 23771897 AGCUAAUAAUUAACAUGUCGAUACUGUCUGCGGAUUGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCU .............(((((((((.((.....(((..((((...(((((((.(.....).)))))))..))))...))).)))))))....))))............. ( -23.50) >DroSec_CAF1 2137 106 - 1 AGCUAAUAAUUAACAUGUCGAUACUGUCUGCGGAUCGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCU ...................((((.((..((((((((((((..(((((((.(.....).)))))))...((......)).))))))...)).))))..))..)))). ( -23.30) >DroSim_CAF1 2141 106 - 1 AGCUAAUAAUUAACAUGUCGAUACUGUCCGCGGAUCGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCU .....((((((...((((.(((.(((....))).((((((..(((((((.(.....).)))))))...((......)).))))))..)))..))))..)))))).. ( -21.90) >DroEre_CAF1 2083 106 - 1 AGCUAAUAAUUAACAUGUCGAUACUGUCUGCAGUUUGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUAUGGCAGAUCGAACAUCAUGCAUUCAAUUAUCU ...................((((.((..((((((((((((...((((((.(.....).))))))(((((....))))).))))))))....))))..))..)))). ( -26.30) >DroYak_CAF1 2007 106 - 1 AGCUAAUAAUUAACAUGUCGAUACUGUCUGCAGUUUGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUAUGGAAGAUCGAACAUCAUGCACUCAAUUAUCU ...................((((.((..((((((((((((..(((((((.(.....).)))))))((((....))))..))))))))....))))..))..)))). ( -24.90) >consensus AGCUAAUAAUUAACAUGUCGAUACUGUCUGCGGAUUGAUCAUCAUCGAUGGACACGCGAUCGAUGUCAUCAUUACCGAAGAUCGAACAUCAUGCAUUCAAUUAUCU .............((((..(...(((....))).((((((..(((((((.(.....).)))))))...((......)).)))))).)..))))............. (-19.82 = -19.54 + -0.28)

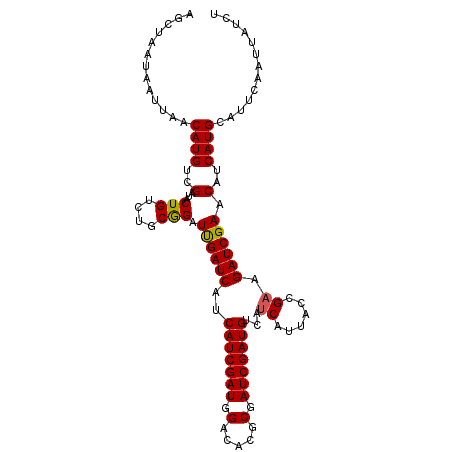

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:58 2006