| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,795,350 – 1,795,443 |
| Length | 93 |
| Max. P | 0.588099 |

| Location | 1,795,350 – 1,795,443 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.13 |
| Mean single sequence MFE | -26.69 |
| Consensus MFE | -12.21 |
| Energy contribution | -15.67 |
| Covariance contribution | 3.46 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
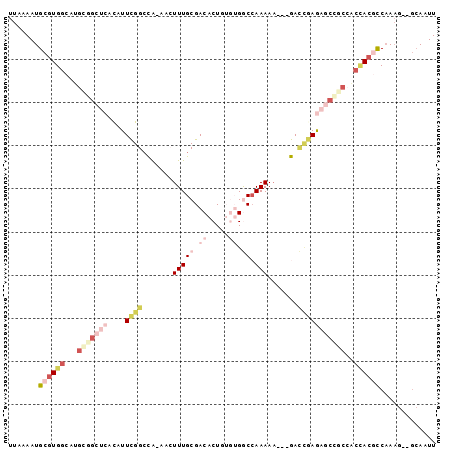
>3L_DroMel_CAF1 1795350 93 + 23771897 UUAAAAUGCGUGGCAUGCGGCUCACAUUCGGCCA-AACUUUGCGACACUGUGUGGCCAAAAA---GACCGAGAGCUGCCACCACGCCAAAG--GCAAUU .......((((((...(((((((.(..(((((((-.((..((...))..)).))))).....---))..).)))))))..)))))).....--...... ( -30.70) >DroVir_CAF1 11038 89 + 1 UUAAAAUGCGUGGCAUGCG-CGCACAUUUGGCUG-CA-UUUGCGACACUGAAAGACAAAAAAAA-AAA--AAAA--ACAACUAAAUCCAAGCC--AAUU ......((((((.....))-))))...(((((((-(.-...)).....((.....)).......-...--....--.............))))--)).. ( -15.20) >DroSec_CAF1 10810 93 + 1 UUAAAAUGCGUGGCAUGCGGCUCACAUUCGGGCA-AACUUUGCGACACUGUGUGGCCAAAAA---GACCGAGAGCCGCCGCCACGCCAAAG--GCAAUU .......(((((((..(((((((....((((...-......((.((.....)).))......---..))))))))))).))))))).....--...... ( -36.73) >DroSim_CAF1 10819 93 + 1 UUAAAAUGCGUGGCAUGCGGCUCACAUUCGGCCA-AACUUUGCGACACUGUGUGGCCAAAAA---GACCGAGAGCCGCCGCCACGCCAAAG--GCAAUU .......(((((((..(((((((.(..(((((((-.((..((...))..)).))))).....---))..).))))))).))))))).....--...... ( -36.40) >DroEre_CAF1 10938 93 + 1 UUAAAAUGCGUGGCAUGUGGCUCGUAUUCGGCCA-AACUUUGCGACACUUUGUGGCCAAAAC---GACUGAGAGCCGCCACCACGCCAAAG--GCAAUU .......((((((...((((((((....)(((((-.....((...)).....))))).....---......)))))))..)))))).....--...... ( -29.90) >DroWil_CAF1 15244 88 + 1 UUAAAAUGCGUGGCAUGCGGCACACAUUUGGCUACAAAUUUGCGACACUGAAAGACCAAAAAAUAAAUUAAAA---------ACCCCAACG--GCAAUU ......(((((.(((((..((.((....))))..))....))).))..((......))...............---------.........--)))... ( -11.20) >consensus UUAAAAUGCGUGGCAUGCGGCUCACAUUCGGCCA_AACUUUGCGACACUGUGUGGCCAAAAA___GACCGAGAGCCGCCACCACGCCAAAG__GCAAUU .......((((((...(((((((....((((.......(((((.((.....)).)).))).......)))))))))))..))))))............. (-12.21 = -15.67 + 3.46)


| Location | 1,795,350 – 1,795,443 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.13 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -14.56 |
| Energy contribution | -17.19 |
| Covariance contribution | 2.63 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
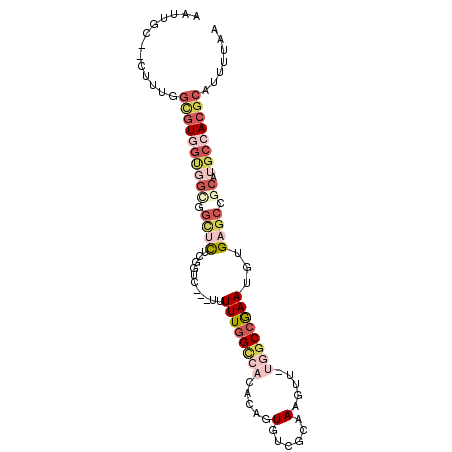
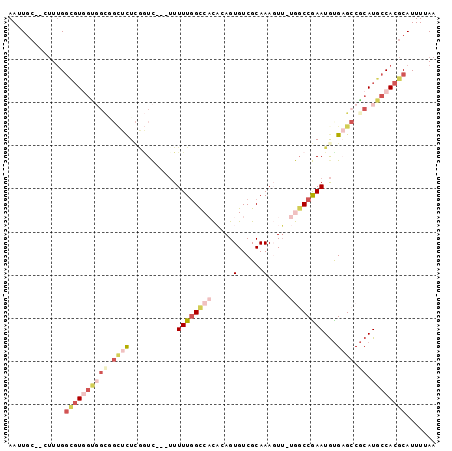
>3L_DroMel_CAF1 1795350 93 - 23771897 AAUUGC--CUUUGGCGUGGUGGCAGCUCUCGGUC---UUUUUGGCCACACAGUGUCGCAAAGUU-UGGCCGAAUGUGAGCCGCAUGCCACGCAUUUUAA ......--.....((((((((((.((((.((...---..((((((((.((..((...))..)).-)))))))))).)))).)).))))))))....... ( -35.00) >DroVir_CAF1 11038 89 - 1 AAUU--GGCUUGGAUUUAGUUGU--UUUU--UUU-UUUUUUUUGUCUUUCAGUGUCGCAAA-UG-CAGCCAAAUGUGCG-CGCAUGCCACGCAUUUUAA ...(--(((..((((..((....--....--...-....))..))))....(((.((((.(-(.-.......)).))))-)))..)))).......... ( -13.46) >DroSec_CAF1 10810 93 - 1 AAUUGC--CUUUGGCGUGGCGGCGGCUCUCGGUC---UUUUUGGCCACACAGUGUCGCAAAGUU-UGCCCGAAUGUGAGCCGCAUGCCACGCAUUUUAA ......--.....(((((((((((((((..((((---.....))))..(((.....(((.....-))).....)))))))))).))))))))....... ( -40.40) >DroSim_CAF1 10819 93 - 1 AAUUGC--CUUUGGCGUGGCGGCGGCUCUCGGUC---UUUUUGGCCACACAGUGUCGCAAAGUU-UGGCCGAAUGUGAGCCGCAUGCCACGCAUUUUAA ......--.....(((((((((((((((.((...---..((((((((.((..((...))..)).-)))))))))).))))))).))))))))....... ( -43.70) >DroEre_CAF1 10938 93 - 1 AAUUGC--CUUUGGCGUGGUGGCGGCUCUCAGUC---GUUUUGGCCACAAAGUGUCGCAAAGUU-UGGCCGAAUACGAGCCACAUGCCACGCAUUUUAA ...(((--...(((((((.(((((((.....)))---((((((((((.((..(......)..))-)))))))).))..))))))))))).)))...... ( -33.40) >DroWil_CAF1 15244 88 - 1 AAUUGC--CGUUGGGGU---------UUUUAAUUUAUUUUUUGGUCUUUCAGUGUCGCAAAUUUGUAGCCAAAUGUGUGCCGCAUGCCACGCAUUUUAA ...(((--.((.((.((---------...............(((....)))(((.((((.(((((....))))).)))).))))).)))))))...... ( -15.60) >consensus AAUUGC__CUUUGGCGUGGUGGCGGCUCUCGGUC___UUUUUGGCCACACAGUGUCGCAAAGUU_UGGCCGAAUGUGAGCCGCAUGCCACGCAUUUUAA .............((((((((((.((((...........((((((((.....(......).....))))))))...)))).)).))))))))....... (-14.56 = -17.19 + 2.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:51 2006