
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,312,133 – 16,312,333 |
| Length | 200 |
| Max. P | 0.953367 |

| Location | 16,312,133 – 16,312,253 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -44.37 |
| Consensus MFE | -30.83 |
| Energy contribution | -31.00 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16312133 120 + 23771897 AGGGAUCCGGUGGCUAUAGCAUUGGCAGCGGCCUGCGUUCCAUUGCCCAGGGAUCGGCGGAUCAGGCUCACAGUGCAGUGACCAACCAGCAUGCAGCUGCCAAGCAGGCGGCCUAUAUAG ........(((.(((...((.((((((((.((.((((.(((...(((........)))))).).((.((((......)))))).....))).)).)))))))))).))).)))....... ( -46.00) >DroVir_CAF1 25727 114 + 1 AGC------CUGGCUAUAGCAUAGGCAGCGGCUUGCGCUCCAUAGCCCAGGGAUCAGCGGAUCAGGCGUACACAGCCGUGGCCAAUCAGCAUGCGGCGGCCAAGCAGGCCGCCUUCAUAG .((------(((((....)).))))).(((((((((...((...(((....((((....)))).))).......((((((((......)).))))))))....)))))))))........ ( -48.90) >DroPse_CAF1 9589 120 + 1 AGGGGAAUCAUGGCUACAGCAUUGGCAGCGGACUGCGUUCCAUAGCCCAGGGAUCAGCGGAUCAGGCGCACAGCGCCGUGUCCAACCAGCAUGCGGCGGCCAAGCAAGCGGCAUACAUUG ............((..(.((.(((((.((.(..((((((((........)))))....((((..(((((...)))))..)))).....)))..).)).)))))))..)..))........ ( -39.00) >DroGri_CAF1 19703 114 + 1 AGU------CUGGCUAUAGCAUUGGCAGUGGUCUGCGCUCCAUAGCACAAGGAUCCGCGGAUCAGGCGCACAGCGCUGUGGCCAAUCAACAUGCGGCAGCCAAACAAGCUGCAUUUAUCG .(.------.((((((((((.(((((..(((((((((.(((.........)))..)))))))))...)).))).))))))))))..)...(((((((..........)))))))...... ( -45.40) >DroMoj_CAF1 22344 114 + 1 AGG------CAGGUUACAGCAUAGGCAGCGGCCUGCGCUCCAUUGCCCAAGGAUCGGCAGAUCAGGCUUACAGCGCCGUCGCAAAUCAACAUGCUGCCGCUAAGCAGGCUGCUUUUAUAG ..(------(........))(((((.(((((((((((((.....(((....((((....)))).)))....)))((.((.(((........))).)).))...))))))))))))))).. ( -40.80) >DroAna_CAF1 31755 120 + 1 AGGGAAGUGCUGGCUACAGCAUAGGCAGUGGCCUGCGCUCUAUCGCUCAGGGAUCAGCGGAUCAGGCUUAUAGUGCUGUGGCUAACCAACAUGCUGCUGCCAAGCAGGCGGCUUACAUAG ......((..((((((((((((((((.....((((.((......)).))))((((....))))..))))...))))))))))))....))(((..((((((.....))))))...))).. ( -46.10) >consensus AGG______CUGGCUACAGCAUAGGCAGCGGCCUGCGCUCCAUAGCCCAGGGAUCAGCGGAUCAGGCGCACAGCGCCGUGGCCAACCAACAUGCGGCGGCCAAGCAGGCGGCAUACAUAG ..........(((((((.((...((.((((.....))))))...(((....((((....)))).))).......)).)))))))...........((((((.....))))))........ (-30.83 = -31.00 + 0.17)


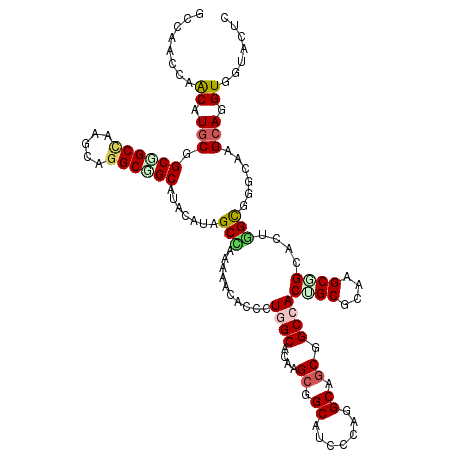
| Location | 16,312,213 – 16,312,333 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -49.78 |
| Consensus MFE | -25.07 |
| Energy contribution | -25.35 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16312213 120 + 23771897 ACCAACCAGCAUGCAGCUGCCAAGCAGGCGGCCUAUAUAGCCCAGAAUACACUGGCCCAAGCGGCUUCUCAGGCGGCGGCCACUGCACAAGCGGCUCUGGUGGGCAAGCAGGUUGUUCUC ..(((((.((.(((.((((((.....))))))........((((((......(((((...((.(((.....))).)))))))((((....)))).))))).).))).)).)))))..... ( -48.40) >DroVir_CAF1 25801 120 + 1 GCCAAUCAGCAUGCGGCGGCCAAGCAGGCCGCCUUCAUAGCCAAAAACACGCUGGCACAGGCCGCAUCGCAGGCCGCCGCAACGGCGCAGGCAGCACUGGCCGGCAAGCAGGUCAUACUC (((........((((((((((..((..((.((((.....((((.........))))..)))).))...)).))))))))))..(((.(((......)))))))))............... ( -52.90) >DroGri_CAF1 19777 120 + 1 GCCAAUCAACAUGCGGCAGCCAAACAAGCUGCAUUUAUCGCUAAAAAUACAUUGGCACAGGCUGCCUCUCAGGCAGCGGCCACUGCGCAAGCAGCACUGGCUGGCAAGCAAGUGUUGCUG ...........(((((((((.......))))).................((.((((....((((((.....)))))).)))).))))))(((((((((.(((....))).))))))))). ( -49.30) >DroMoj_CAF1 22418 120 + 1 GCAAAUCAACAUGCUGCCGCUAAGCAGGCUGCUUUUAUAGCCAAAAACACCUUGGCGCAAGCAGCAUCACAGGCAGCUGCCACAGCGCAAGCGGCGCUCGCGGGCAAGCAGGUGAUACUA ....((((...((((((((((..((..(((((((.....(((((.......)))))..)))))))......(((....)))...))...))))))(((....))).))))..)))).... ( -48.30) >DroAna_CAF1 31835 120 + 1 GCUAACCAACAUGCUGCUGCCAAGCAGGCGGCUUACAUAGCCCAAAACACCCUGGCUCAGGCGGCAUCCCAAGCGGCCGCCACUGCGCAGGCGGCCUUGGUGGGAAAGCAGGUAGUUCUC ((((.((....((((((((((.((((((.((((.....))))........))).)))..)))))).(((((...(((((((........)))))))....))))).)))))))))).... ( -51.20) >DroPer_CAF1 5581 120 + 1 UCCAACCAGCAUGCGGCGGCCAAGCAGGCGGCAUACAUUGCACAGAACACCCUCGCGCAAGCGGCAUCCCAGGCAUCGGCCACGGCCCAGGCUGCGCUUGCGGGGAAGGAGGUGGUGCUG ......((((((...((.(((.....))).))...............(((((((.((((((((........(((....))).((((....)))))))))))).)...)).)))))))))) ( -48.60) >consensus GCCAACCAACAUGCGGCGGCCAAGCAGGCGGCAUACAUAGCCAAAAACACCCUGGCACAAGCGGCAUCCCAGGCAGCGGCCACUGCGCAAGCGGCACUGGCGGGCAAGCAGGUGGUACUC ........((.(((.((((((.....)))))).......(((..........((((....((.((.......)).)).))))((((....))))....)))......))).))....... (-25.07 = -25.35 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:32 2006