
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,293,158 – 16,293,275 |
| Length | 117 |
| Max. P | 0.596381 |

| Location | 16,293,158 – 16,293,275 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -45.83 |
| Consensus MFE | -30.50 |
| Energy contribution | -31.76 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
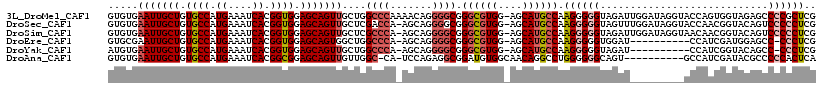
>3L_DroMel_CAF1 16293158 117 + 23771897 GUGUGAAUUGCUGUGCCAUGAAAUCACGGUGGAGCAGUUGCUGGCCCAAAACAGGGGCGGGCGUGG-AGCAUGCCAAGGGGGUAGAUUGGAUAGGUACCAGUGGUAGAGCCCCGCUCG .....(((((((.((((.((....)).)))).)))))))(((.((((.......)))).)))((((-.((.(((((....((((..(.....)..))))..)))))..)).))))... ( -46.80) >DroSec_CAF1 5322 116 + 1 GUGUGAAUUGCUGUGCCAUGAAAUCACGGUGGAGCAGUUGCUCGACCA-AGCAGGGGCGGGCGUGG-AGCAUGCCAAGGGGGUAGUUUGGAUAGGUACCAACGGUACAGUCCCCCUCG ((((((((((((.((((.((....)).)))).)))))))(((((.((.-.....)).)))))....-..)))))...(((((......((((..(((((...))))).))))))))). ( -43.40) >DroSim_CAF1 5632 116 + 1 GUGUGAAUUGCUGUGCCAUGAAAUCACGGUGGAGCAGUUGCUCGCCCA-AGCAGGGGCGGGCGUGG-AGCAUGCCAAGGGGGUAGAUUGGAUAGGUAACAACGGUACAGUCCCCCUCG ((((((((((((.((((.((....)).)))).)))))))((((((((.-.....))))))))....-..)))))...(((((..(((((....(.......)....))))).))))). ( -47.30) >DroEre_CAF1 6711 105 + 1 GUGCGAAUUGCUGUGCCAUGAAAUCACGGUGGAGCAGUGGCUGGCCCA-AGCAGGGGCGGGCGUGG-AGCAUGCCAAGGGGGUGGAU----------CCAUCGAUGGAGCC-CCCUCG ((((..((((((.((((.((....)).)))).)))))).(((.((((.-.....)))).)))....-.)))).....(((((.((.(----------(((....)))).))-))))). ( -48.40) >DroYak_CAF1 12579 105 + 1 AUGUGAAUUGCUGUGCCAUGAAAUCACGGUGGAGCAGUUGCUGGCCCA-AGCAGGGGCGGGCGUGG-AGCAUGCCAAGGGGGUAGAU----------CCAUCGGUACAGCC-CCCUCG ((((.(((((((.((((.((....)).)))).)))))))(((.((((.-.....)))).)))....-.))))....(((((((.(((----------(....))).).)))-)))).. ( -43.90) >DroAna_CAF1 7571 106 + 1 GUGUGAAUUGCUGUGCCAUGAAAUCACGGCGGAGCAGUUGUUGGC-CA-UCCAGAGGCGGAUGUGGCAACAGGCCUGGGGGGCAGU----------GCCAUCGAUACGCCCCCACUCA .....(((((((.((((.((....)).)))).)))))))...(((-((-(((......)))).((....))))))..((((((.((----------(.......)))))))))..... ( -45.20) >consensus GUGUGAAUUGCUGUGCCAUGAAAUCACGGUGGAGCAGUUGCUGGCCCA_AGCAGGGGCGGGCGUGG_AGCAUGCCAAGGGGGUAGAU_________ACCAUCGGUACAGCCCCCCUCG .....(((((((.((((.((....)).)))).)))))))....((((.......)))).((((((....)))))).((((((............................)))))).. (-30.50 = -31.76 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:26 2006