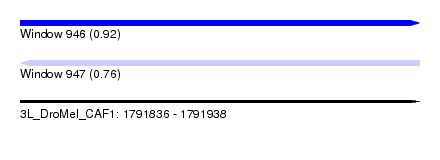
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,791,836 – 1,791,938 |
| Length | 102 |
| Max. P | 0.921798 |

| Location | 1,791,836 – 1,791,938 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -38.49 |
| Consensus MFE | -24.29 |
| Energy contribution | -23.35 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
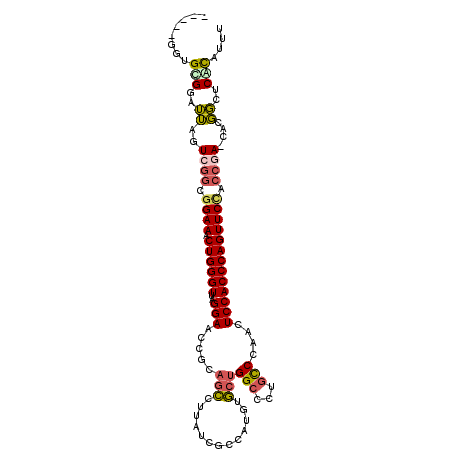
>3L_DroMel_CAF1 1791836 102 + 23771897 AAAUGUGAGCCGUG-UCGGUGGAACUGGGUGGAGUUGGGCAGGGCCAGCACAUGGCGAUAAGGCUGCGGUUCCUAACCCAGUUUCCGCCGACUAAUCCGCACA----- ...((((......(-(((((((((((((((((((..((((...((((.....))))......))).)..))))..)))))).)))))))))).......))))----- ( -46.22) >DroSec_CAF1 7305 102 + 1 AAAUGCGAGCCGUG-UCGGUGGAACUGGGUGGAGUUGGGCAGGGCCAGCACAUGCCGAUAAGGCUGCGGUUCCUAACCCAGUUUCCGCCGACUAAUCCGCACA----- ...((((.(....(-(((((((((((((((((((((((......))))).((.(((.....)))))....)))..)))))).))))))))))....)))))..----- ( -47.60) >DroSim_CAF1 7323 102 + 1 AAAUGCGAGCCGUG-UCGGUGGAACUGGGUGGAGUUGGGCAGGGCCAGCACAUGCCGAUAAGGCUGCGGUUCCUAACCCAGUUUCCGCCGACUAAUCCGCACG----- ...((((.(....(-(((((((((((((((((((((((......))))).((.(((.....)))))....)))..)))))).))))))))))....)))))..----- ( -47.60) >DroEre_CAF1 7516 103 + 1 AAAUGCGAGCCGUA-UAGGUAGAACUGGGUGGAGUUGGGCAGGGCCAGCAUAUAGCGAUAAGGCUUCGGUUCCUAACCCAGUUUCCGCCGACUAAACCACUCCA---- ..(((((...))))-).(((.(((((((((((((..((....((((.((.....)).....))))))..))))..)))))))))..)))...............---- ( -30.40) >DroYak_CAF1 7582 107 + 1 AAAUGUGAGCCGUG-UCGGUAGAACUGGGUGGAGUUGGGCAGGGCCAGCACAUAGCGAUAAGGCUUCGUUUCCUAACCCAGUUUCCGCCGACUAAUCCACUCCAAUCC ...((.(((..(.(-(((((.(((((((((((((((((......)))))....(((((.......))))))))..)))))))))..))))))....)..))))).... ( -38.10) >DroPer_CAF1 7093 91 + 1 AAAAAUGAGAAACGAUCUUCAGAACUGGGUGGAGUUUGAUU---CCAAUACA----GAUACGAG--UAGAUCCCAACCCAGUUUCAAUCCAAUCUCCCGC-------- ......((((...(((.....(((((((((((..(((.(((---(.......----.....)))--))))..)).)))))))))..)))...))))....-------- ( -21.00) >consensus AAAUGCGAGCCGUG_UCGGUAGAACUGGGUGGAGUUGGGCAGGGCCAGCACAUGGCGAUAAGGCUGCGGUUCCUAACCCAGUUUCCGCCGACUAAUCCGCACA_____ ....(((.((((....)))).(((((((((((((((((......)))))......((.........))..)))..))))))))).))).................... (-24.29 = -23.35 + -0.94)

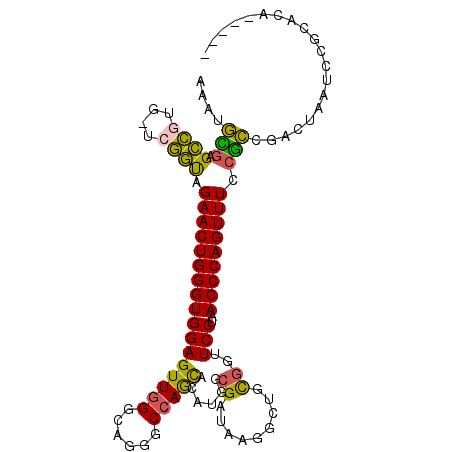
| Location | 1,791,836 – 1,791,938 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1791836 102 - 23771897 -----UGUGCGGAUUAGUCGGCGGAAACUGGGUUAGGAACCGCAGCCUUAUCGCCAUGUGCUGGCCCUGCCCAACUCCACCCAGUUCCACCGA-CACGGCUCACAUUU -----((((.((....(((((.((((.((((((..(((...((((.......((((.....)))).)))).....))))))))))))).))))-)....))))))... ( -38.00) >DroSec_CAF1 7305 102 - 1 -----UGUGCGGAUUAGUCGGCGGAAACUGGGUUAGGAACCGCAGCCUUAUCGGCAUGUGCUGGCCCUGCCCAACUCCACCCAGUUCCACCGA-CACGGCUCGCAUUU -----.(((((..((.(((((.((((.((((((..(((..(((((((.....))).))))..(((...)))....))))))))))))).))))-)..))..))))).. ( -43.70) >DroSim_CAF1 7323 102 - 1 -----CGUGCGGAUUAGUCGGCGGAAACUGGGUUAGGAACCGCAGCCUUAUCGGCAUGUGCUGGCCCUGCCCAACUCCACCCAGUUCCACCGA-CACGGCUCGCAUUU -----.(((((..((.(((((.((((.((((((..(((..(((((((.....))).))))..(((...)))....))))))))))))).))))-)..))..))))).. ( -43.80) >DroEre_CAF1 7516 103 - 1 ----UGGAGUGGUUUAGUCGGCGGAAACUGGGUUAGGAACCGAAGCCUUAUCGCUAUAUGCUGGCCCUGCCCAACUCCACCCAGUUCUACCUA-UACGGCUCGCAUUU ----..((((.((.((...((..(((.((((((..(((.....(((......))).......(((...)))....))))))))))))..))))-.)).))))...... ( -29.10) >DroYak_CAF1 7582 107 - 1 GGAUUGGAGUGGAUUAGUCGGCGGAAACUGGGUUAGGAAACGAAGCCUUAUCGCUAUGUGCUGGCCCUGCCCAACUCCACCCAGUUCUACCGA-CACGGCUCACAUUU (((((((.(((((......((((....).(((((((...(((.(((......))).))).))))))).)))....))))))))))))..(((.-..)))......... ( -37.20) >DroPer_CAF1 7093 91 - 1 --------GCGGGAGAUUGGAUUGAAACUGGGUUGGGAUCUA--CUCGUAUC----UGUAUUGG---AAUCAAACUCCACCCAGUUCUGAAGAUCGUUUCUCAUUUUU --------..(((((((..((((..((((((((.(((...((--(.......----.)))(((.---...))).))).)))))))).....)))))))))))...... ( -23.70) >consensus _____GGUGCGGAUUAGUCGGCGGAAACUGGGUUAGGAACCGCAGCCUUAUCGCCAUGUGCUGGCCCUGCCCAACUCCACCCAGUUCCACCGA_CACGGCUCACAUUU ........(((..((..((((.((((.((((((..(((.....(((.............)))(((...)))....))))))))))))).))))....))..))).... (-22.10 = -22.30 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:50 2006