
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,266,464 – 16,266,597 |
| Length | 133 |
| Max. P | 0.982242 |

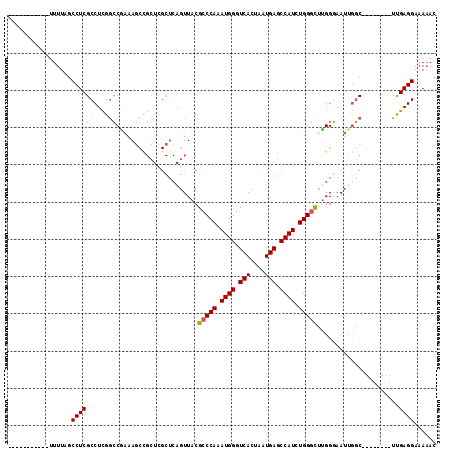
| Location | 16,266,464 – 16,266,560 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -33.46 |
| Consensus MFE | -22.78 |
| Energy contribution | -22.86 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16266464 96 + 23771897 -----------CUUUAGCCUCGCCUCGGCCGAAAGCCGCUCGCUCAGUUACGCCCAAAUGGGUCACUAAUGAGCCAUCUGGGCCUGGGAAUUGGC--------UUGAGGAAAAAC -----------......((((((....))...((((((....(((((....(((((.((((.(((....))).)))).))))))))))...))))--------))))))...... ( -38.40) >DroSec_CAF1 17805 84 + 1 -----------UUUUAGCCUCGCCUCGGCCGAAA------------GUUACGCCCAAAUGGGUCACUAAUGAGCCAUCUGGGCUUGGGAAUUGGC--------UUGAGGAAAAAC -----------...........((((((((((..------------.(((.(((((.((((.(((....))).)))).))))).)))...)))))--------.)))))...... ( -32.10) >DroSim_CAF1 16337 96 + 1 -----------UUUUAGCCUCGCCUCGGCCGAAAGCCGCUCGCUCAGUUACGCCCAAAUGGGUCACUAAUGAGCCAUCUGGGCUUGGGAAUUGGC--------UUGAGGAAAAAC -----------......((((((....))...((((((....(((((....(((((.((((.(((....))).)))).))))))))))...))))--------))))))...... ( -36.40) >DroEre_CAF1 17786 105 + 1 UUCAGCCACAGUUUUAGCCUCGCCUCGACCA-AAGCCGCUCGCUCAGUUACGCCCAAAUGGGUCACUAAUGAGCCAUCUGGGUCUGGGAAUUGGC--------UUGAGGAAA-AC ......................(((((....-.(((((....(((((....(((((.((((.(((....))).)))).))))))))))...))))--------))))))...-.. ( -33.10) >DroAna_CAF1 16687 109 + 1 -----CUUGGCUUUCAGCCUCGCCUUGGUCCAAA-CUGCUCGCUCAGUUACGCCCAAAUGGGUCACUAAUGAGCCAUCUGGUCUGGGAAAUACACAAACAUAGUCGAGGAAAAAC -----(((((((....((...))((..(.(((..-..(((((...(((...((((....)))).)))..)))))....))).)..))..............)))))))....... ( -27.30) >consensus ___________UUUUAGCCUCGCCUCGGCCGAAAGCCGCUCGCUCAGUUACGCCCAAAUGGGUCACUAAUGAGCCAUCUGGGCUUGGGAAUUGGC________UUGAGGAAAAAC .................((((((....))......................(((((.((((.(((....))).)))).)))))......................))))...... (-22.78 = -22.86 + 0.08)


| Location | 16,266,464 – 16,266,560 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -26.28 |
| Energy contribution | -27.46 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16266464 96 - 23771897 GUUUUUCCUCAA--------GCCAAUUCCCAGGCCCAGAUGGCUCAUUAGUGACCCAUUUGGGCGUAACUGAGCGAGCGGCUUUCGGCCGAGGCGAGGCUAAAG----------- .(((..((((..--------(((..(((((((((((((((((.(((....))).))))))))))....))).).)))((((.....)))).)))))))..))).----------- ( -38.60) >DroSec_CAF1 17805 84 - 1 GUUUUUCCUCAA--------GCCAAUUCCCAAGCCCAGAUGGCUCAUUAGUGACCCAUUUGGGCGUAAC------------UUUCGGCCGAGGCGAGGCUAAAA----------- (((((.((((..--------(((.........((((((((((.(((....))).)))))))))).....------------....))).)))).))))).....----------- ( -34.07) >DroSim_CAF1 16337 96 - 1 GUUUUUCCUCAA--------GCCAAUUCCCAAGCCCAGAUGGCUCAUUAGUGACCCAUUUGGGCGUAACUGAGCGAGCGGCUUUCGGCCGAGGCGAGGCUAAAA----------- (((((.((((..--------(((.........((((((((((.(((....))).))))))))))......((((.....))))..))).)))).))))).....----------- ( -38.00) >DroEre_CAF1 17786 105 - 1 GU-UUUCCUCAA--------GCCAAUUCCCAGACCCAGAUGGCUCAUUAGUGACCCAUUUGGGCGUAACUGAGCGAGCGGCUU-UGGUCGAGGCGAGGCUAAAACUGUGGCUGAA ..-((.((((..--------(((((..(((...(((((((((.(((....))).)))))))))(((......))).).))..)-)))).)))).))(((((......)))))... ( -35.20) >DroAna_CAF1 16687 109 - 1 GUUUUUCCUCGACUAUGUUUGUGUAUUUCCCAGACCAGAUGGCUCAUUAGUGACCCAUUUGGGCGUAACUGAGCGAGCAG-UUUGGACCAAGGCGAGGCUGAAAGCCAAG----- (((((((((((.((.((((((.(.....).))))((((((.((((.(((((..(((....)))....)))))..)))).)-)))))..)).)))))))..))))))....----- ( -34.40) >consensus GUUUUUCCUCAA________GCCAAUUCCCAAGCCCAGAUGGCUCAUUAGUGACCCAUUUGGGCGUAACUGAGCGAGCGGCUUUCGGCCGAGGCGAGGCUAAAA___________ (((((.((((......................((((((((((.(((....))).))))))))))((....((((.....))))...)).)))).)))))................ (-26.28 = -27.46 + 1.18)



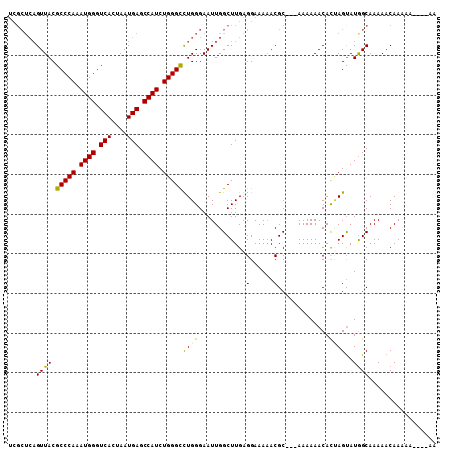
| Location | 16,266,492 – 16,266,597 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -19.30 |
| Energy contribution | -18.61 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16266492 105 + 23771897 UCGCUCAGUUACGCCCAAAUGGGUCACUAAUGAGCCAUCUGGGCCUGGGAAUUGGCUUGAGGAAAAACGC---AAAAAACAUCAGUAUGGCAAAAACAAAAAAAAAAA ...(((((....(((((.((((.(((....))).)))).)))))))))).....(((((.(......).)---))....(((....)))))................. ( -24.00) >DroSec_CAF1 17828 95 + 1 -------GUUACGCCCAAAUGGGUCACUAAUGAGCCAUCUGGGCUUGGGAAUUGGCUUGAGGAAAAACGC---AAAAAACACUAGUAUGGCAAAAACAAAAAA---AA -------((((.(((((.((((.(((....))).)))).)))))(..((......))..)..........---..............))))............---.. ( -21.10) >DroSim_CAF1 16365 100 + 1 UCGCUCAGUUACGCCCAAAUGGGUCACUAAUGAGCCAUCUGGGCUUGGGAAUUGGCUUGAGGAAAAACGC---AAAAAACACUAGUAUGGCAAAAACAAAAA-----A ..(((....((((((((.((((.(((....))).)))).)))))(..((......))..)..........---...........))).)))...........-----. ( -24.00) >DroEre_CAF1 17824 100 + 1 UCGCUCAGUUACGCCCAAAUGGGUCACUAAUGAGCCAUCUGGGUCUGGGAAUUGGCUUGAGGAAA-ACGCAACAAAAAACACAAGCAUAGCAAAA-CAAA------AA ..((((((((..(((((.((((.(((....))).)))).))))).....)))))(((((.(....-.............).)))))..)))....-....------.. ( -26.23) >consensus UCGCUCAGUUACGCCCAAAUGGGUCACUAAUGAGCCAUCUGGGCCUGGGAAUUGGCUUGAGGAAAAACGC___AAAAAACACUAGUAUGGCAAAAACAAAAA____AA .......((((.(((((.((((.(((....))).)))).)))))((((............(......).............))))..))))................. (-19.30 = -18.61 + -0.69)


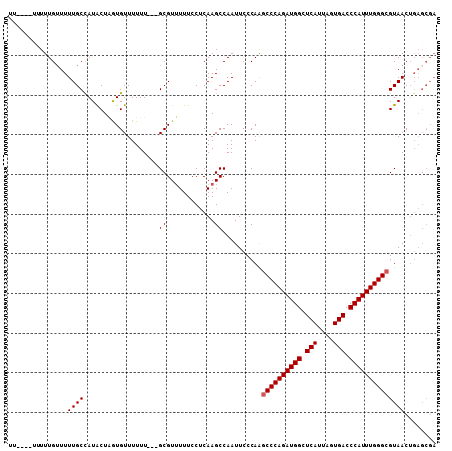
| Location | 16,266,492 – 16,266,597 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -19.95 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16266492 105 - 23771897 UUUUUUUUUUUGUUUUUGCCAUACUGAUGUUUUUU---GCGUUUUUCCUCAAGCCAAUUCCCAGGCCCAGAUGGCUCAUUAGUGACCCAUUUGGGCGUAACUGAGCGA .........((((((((((....(((.......((---(.((((......)))))))....)))((((((((((.(((....))).))))))))))))))..)))))) ( -27.60) >DroSec_CAF1 17828 95 - 1 UU---UUUUUUGUUUUUGCCAUACUAGUGUUUUUU---GCGUUUUUCCUCAAGCCAAUUCCCAAGCCCAGAUGGCUCAUUAGUGACCCAUUUGGGCGUAAC------- ..---..........((((..............((---(.((((......))))))).......((((((((((.(((....))).)))))))))))))).------- ( -23.40) >DroSim_CAF1 16365 100 - 1 U-----UUUUUGUUUUUGCCAUACUAGUGUUUUUU---GCGUUUUUCCUCAAGCCAAUUCCCAAGCCCAGAUGGCUCAUUAGUGACCCAUUUGGGCGUAACUGAGCGA .-----.........(((((((((.........((---(.((((......))))))).......((((((((((.(((....))).)))))))))))))..)).)))) ( -26.30) >DroEre_CAF1 17824 100 - 1 UU------UUUG-UUUUGCUAUGCUUGUGUUUUUUGUUGCGU-UUUCCUCAAGCCAAUUCCCAGACCCAGAUGGCUCAUUAGUGACCCAUUUGGGCGUAACUGAGCGA ..------.(((-(((.(.(((((....((((...((((.((-((.....))))))))....))))((((((((.(((....))).))))))))))))).).)))))) ( -26.40) >consensus UU____UUUUUGUUUUUGCCAUACUAGUGUUUUUU___GCGUUUUUCCUCAAGCCAAUUCCCAAGCCCAGAUGGCUCAUUAGUGACCCAUUUGGGCGUAACUGAGCGA ...............((((...................((............))..........((((((((((.(((....))).))))))))))))))........ (-19.95 = -20.20 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:22 2006