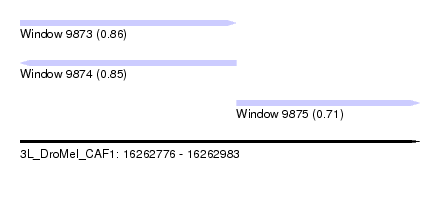
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,262,776 – 16,262,983 |
| Length | 207 |
| Max. P | 0.858625 |

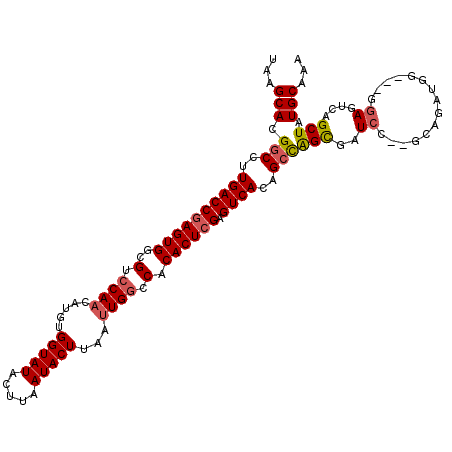
| Location | 16,262,776 – 16,262,888 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -28.14 |
| Energy contribution | -29.18 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16262776 112 + 23771897 UUUGCAUAGCUGACUCCCUCCCAUCUGC--GGAUCGCUGGCUGUGACUCGAGUGUGGCCAAUUAAGUAUUAAGUAUACCACAUGUUGGACGCCACUCGGUCAAGGCCGUGCUUA ...(((((((.((.((((........).--))))))))((((.((((.((((((((.(((((...((((.....)))).....))))).)).)))))))))).))))))))... ( -39.90) >DroSec_CAF1 14192 109 + 1 UUUGCAUAGCUGACUCC---CCAUCUGC--GGAUCGCUGGCUGUGACUCGAGUGUGGCCAAUUAAGUAUUAAGUAUACCACAUGUUGGACGCCACUCGGUCAAGGCCGUGCUUA ...(((((((.((.(((---.(....).--))))))))((((.((((.((((((((.(((((...((((.....)))).....))))).)).)))))))))).))))))))... ( -39.70) >DroSim_CAF1 12696 109 + 1 UUUGCAUAGCUGACUCC---CCAUCUGC--GGAUCGCUGGCUGUGACUCGAGUGUGGCCAAUUAAGUAUUAAGUAUACCACAUGUUGGACGCCACUCGGUCAAGGCCGUGCUUA ...(((((((.((.(((---.(....).--))))))))((((.((((.((((((((.(((((...((((.....)))).....))))).)).)))))))))).))))))))... ( -39.70) >DroEre_CAF1 14168 108 + 1 UUUGCAUAGCUGACUC----CCAUCUGC--GGAUCGCUGGCUGUGACUCGAGUGUGGCCAAUUAAGUAUUAAGUAUACCACAUGUUGGACGCCACUCGGUCAAGGCCGUGCUUA ...(((((((.((.((----((....).--))))))))((((.((((.((((((((.(((((...((((.....)))).....))))).)).)))))))))).))))))))... ( -39.70) >DroAna_CAF1 13482 108 + 1 UUUGCAUAGCGUAUUUU------UUUGAGGUGACUACCAUCUGUGACUCUAGUGUGGCCAAUUAAGUAUUAAGUAUACCACAUGGUGUCCGCCACUCGGUCAAGGUCGUGCUUA ...(((..((.......------....(((((.....))))).(((((...(.(((((.(((.....)))..(.(((((....))))).)))))).))))))..))..)))... ( -26.00) >consensus UUUGCAUAGCUGACUCC___CCAUCUGC__GGAUCGCUGGCUGUGACUCGAGUGUGGCCAAUUAAGUAUUAAGUAUACCACAUGUUGGACGCCACUCGGUCAAGGCCGUGCUUA ...((((...............(((......)))....((((.((((.((((((((.(((((...((((.....)))).....))))).)).)))))))))).))))))))... (-28.14 = -29.18 + 1.04)


| Location | 16,262,776 – 16,262,888 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -27.20 |
| Energy contribution | -27.72 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16262776 112 - 23771897 UAAGCACGGCCUUGACCGAGUGGCGUCCAACAUGUGGUAUACUUAAUACUUAAUUGGCCACACUCGAGUCACAGCCAGCGAUCC--GCAGAUGGGAGGGAGUCAGCUAUGCAAA ...(((.(((..((((((((((..(.((((.....(((((.....)))))...)))).).)))))).))))..)))(((..(((--.(........))))....))).)))... ( -36.10) >DroSec_CAF1 14192 109 - 1 UAAGCACGGCCUUGACCGAGUGGCGUCCAACAUGUGGUAUACUUAAUACUUAAUUGGCCACACUCGAGUCACAGCCAGCGAUCC--GCAGAUGG---GGAGUCAGCUAUGCAAA ...(((.(((..((((((((((..(.((((.....(((((.....)))))...)))).).)))))).))))..)))(((..(((--.(....).---)))....))).)))... ( -38.60) >DroSim_CAF1 12696 109 - 1 UAAGCACGGCCUUGACCGAGUGGCGUCCAACAUGUGGUAUACUUAAUACUUAAUUGGCCACACUCGAGUCACAGCCAGCGAUCC--GCAGAUGG---GGAGUCAGCUAUGCAAA ...(((.(((..((((((((((..(.((((.....(((((.....)))))...)))).).)))))).))))..)))(((..(((--.(....).---)))....))).)))... ( -38.60) >DroEre_CAF1 14168 108 - 1 UAAGCACGGCCUUGACCGAGUGGCGUCCAACAUGUGGUAUACUUAAUACUUAAUUGGCCACACUCGAGUCACAGCCAGCGAUCC--GCAGAUGG----GAGUCAGCUAUGCAAA ...(((.(((..((((((((((..(.((((.....(((((.....)))))...)))).).)))))).))))..)))(((..(((--.......)----))....))).)))... ( -36.40) >DroAna_CAF1 13482 108 - 1 UAAGCACGACCUUGACCGAGUGGCGGACACCAUGUGGUAUACUUAAUACUUAAUUGGCCACACUAGAGUCACAGAUGGUAGUCACCUCAAA------AAAAUACGCUAUGCAAA ...((.((........)).(((((((((.(..((((((....(((.....)))...))))))...).)))...((.(((....)))))...------......))))))))... ( -22.10) >consensus UAAGCACGGCCUUGACCGAGUGGCGUCCAACAUGUGGUAUACUUAAUACUUAAUUGGCCACACUCGAGUCACAGCCAGCGAUCC__GCAGAUGG___GGAGUCAGCUAUGCAAA ...(((.(((..((((((((((..(.((((.....(((((.....)))))...)))).).)))))).))))..)))(((..((...............))....))).)))... (-27.20 = -27.72 + 0.52)


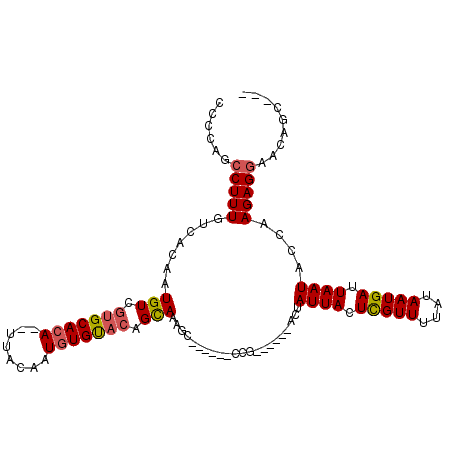
| Location | 16,262,888 – 16,262,983 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.93 |
| Mean single sequence MFE | -21.88 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.46 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16262888 95 + 23771897 CCCCAGCCUUUGUCACAAUGUCGUGCACA--UUACAAUGUGUACAGCAAAGC------CUG------ACUAUUACUCGUUUUAUAAUGAUUAAUACCAAGAGGAACAGC--- ......(((((((((...(((.(((((((--(....)))))))).)))....------.))------))(((((.(((((....))))).)))))...)))))......--- ( -23.40) >DroSec_CAF1 14301 95 + 1 CCCCAGCCUUUGUCACAAUGUCGUGCACA--UUACAAUGUGUACAGCAAAGC------UCG------ACUAUUACUCGUUUUAUAAUGAUUAAUACCAAGAGGAACAGC--- ......((((((((....(((.(((((((--(....)))))))).)))....------..)------))(((((.(((((....))))).)))))...)))))......--- ( -21.80) >DroSim_CAF1 12805 95 + 1 CCCCAGCCUUUGUCACAAUGUCGUGCACA--UUACAAUGUGUACAGCAGAGC------UCG------ACUAUUACUCGUUUUAUAAUGAUUAAUACCAAGAGGAACAGC--- ......((((((((....(((.(((((((--(....)))))))).)))....------..)------))(((((.(((((....))))).)))))...)))))......--- ( -22.90) >DroEre_CAF1 14276 95 + 1 CCCCAGCCUUUGUCACAAUGUCGUGCACA--UUACAAUGUGUACAGCAAAAC------CCG------GCCAUUAUUCGUUUUAUAAUGAUUAAUACCAAGAGGAAUAGC--- ......(((((.......(((.(((((((--(....)))))))).)))....------..(------(..((((.(((((....))))).)))).)).)))))......--- ( -22.70) >DroAna_CAF1 13590 112 + 1 CUUCAGCCUUUGUCACAAUGUCGUGCACAUAUUACACUCUGCCAUGUAAAAUGCGAGUCUGGCUACAAUUAUUAUUUGUUUUUAAAUGAUUAAUUAUAAGAGCAACAACUGC ....((((..((((((......))).)))......((((.((.((.....))))))))..))))...........((((((((((((.....))).)))))))))....... ( -18.60) >consensus CCCCAGCCUUUGUCACAAUGUCGUGCACA__UUACAAUGUGUACAGCAAAGC______CCG______ACUAUUACUCGUUUUAUAAUGAUUAAUACCAAGAGGAACAGC___ ......(((((.......(((.(((((((........))))))).)))......................((((.(((((....))))).))))....)))))......... (-14.14 = -14.46 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:17 2006