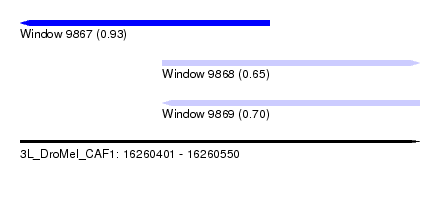
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,260,401 – 16,260,550 |
| Length | 149 |
| Max. P | 0.932134 |

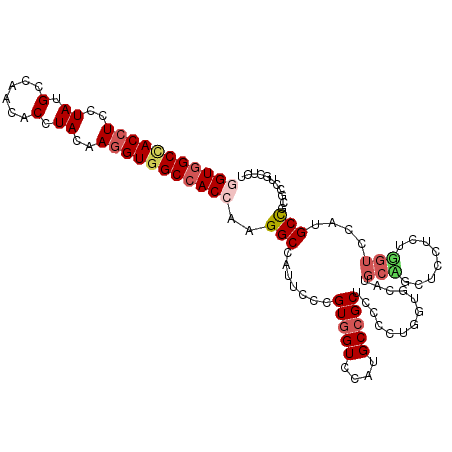
| Location | 16,260,401 – 16,260,494 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16260401 93 - 23771897 GGUGGCCACCUCCUAUGCCAACACCUACAAGGUGGCCACCAAGGCCAUUCCAGUGGUCCAUGCCGCU------------------CCUCUGGUCCAUGCCGUGCCUGCUCU (((((((((((..((.(......).))..))))))))))).(((((.....((((((....))))))------------------.....(((....)))).))))..... ( -37.10) >DroVir_CAF1 11544 108 - 1 GGUGGCCACCUCCUAUGCCAACACGUACAAGGUCGCCACCAAGGCCUAUCCCGUGGUGCAUGCCGCGCCCGUCUAUCAUGCACCCGCCGUUGU---CGCUGCCCCAGCUCU ((((((.((((..((((......))))..)))).))))))..(((..((..((.((((((((..((....))....))))))))))..)).))---)((((...))))... ( -40.40) >DroGri_CAF1 11212 111 - 1 GGUGGCCACAUCCUAUGCCAACACCUACAAGGUGGCGACCAAGGCUUAUCCCGUCGUACAUGCCGCACCCAUCUAUCAUGCACCCGCUGUUGUGCAUGCCGCACCAGCUCU .(((((...............((((.....))))(((((...((.....)).)))))....)))))...................((((..((((.....))))))))... ( -29.50) >DroSec_CAF1 11829 111 - 1 AGUGGCCACCUCCUAUGCCAACACCUACAAGGUGGCCACCAAGGCCAUUCCAGUGGUCCAUGCCGCUCCCCUGGUGCACGCUGCUCCUCUGGUCCAUGCCGUGCCUGCUCU .((((((((((..((.(......).))..))))))))))...(((....((((.((..((.((.((.((...)).))..))))..)).)))).....)))........... ( -39.10) >DroEre_CAF1 11711 111 - 1 GGUGGCCACCUCCUACGCCAACACCUACAAGGUGGCCACCAAGGCCAUCCCCGUGGUCCAUGCCGCUCCCCUGGUGCACGCCGCUCCUCUGGUCCAUGCCGUGCCUGCUCU (((((((((((..((.(......).))..))))))))))).((((.((...(((((.(((.((((......))))((.....)).....))).)))))..))))))..... ( -41.70) >DroAna_CAF1 10995 111 - 1 CGUGGCUACCUCCUACGCCAACACCUACAAGGUGGCCACCAAGGCCAUCCCUGUGGUACACGCCGCCCCUCUGGUCCAUGCUGCUCCUCUGGUCCAUGCUGCCCCUGCUCU .((((((((((..((.(......).))..))))))))))...(((((.....(..(((...((((......))))...)))..).....)))))...((.......))... ( -31.80) >consensus GGUGGCCACCUCCUAUGCCAACACCUACAAGGUGGCCACCAAGGCCAUUCCCGUGGUCCAUGCCGCUCCCCUGGUGCAUGCAGCUCCUCUGGUCCAUGCCGCGCCUGCUCU (((((((((((..((.(......).))..)))))))))))..(((.......(((((....))))).............(((........)))....)))........... (-25.12 = -25.90 + 0.78)

| Location | 16,260,454 – 16,260,550 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.05 |
| Mean single sequence MFE | -41.18 |
| Consensus MFE | -29.45 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16260454 96 + 23771897 GGUGGCCACCUUGUAGGUGUUGGCAUAGGAGGUGGCCACCGGGUGGGC---------------------GA---CAAUGGGAGCGGCGUAGCUCUUGAUGAUCGGGGCGUGCGAGAUCAC ((((((((((((.((.((....)).)).))))))))))))..((((((---------------------..---(....)..))..(((((((((........))))).))))...)))) ( -41.60) >DroVir_CAF1 11612 117 + 1 GGUGGCGACCUUGUACGUGUUGGCAUAGGAGGUGGCCACCGGGUGGAUGGUCUUCACGAUGGGCGCAUGCA---CAAUGGGCGCGGCAUAGCUCUUGAUGAUCGGCGCAUGCGAGAUGAC ((((((.(((((.((.((....)).)).))))).)))))).........(((.((.((....((((...((---(((.((((........))))))).))....))))...)).)).))) ( -45.20) >DroGri_CAF1 11283 117 + 1 GGUCGCCACCUUGUAGGUGUUGGCAUAGGAUGUGGCCACCGGGUGGAUGGUCUUCACGAUGGGUGCAUGGA---CAAUGGGCGCGGCAUAGCUCUUGAUGAUCGGCGCAUGCGAGAUGAC ......((.((((((.((((((((((((((.((.(((.(((.(((((.....)))))..)))((.(((...---..))).))..)))...)))))).))).))))))).)))))).)).. ( -38.50) >DroSec_CAF1 11900 96 + 1 GGUGGCCACCUUGUAGGUGUUGGCAUAGGAGGUGGCCACUGGGUGGGC---------------------GA---CAAUGGGUGCAGCGUAGCUCUUGAUGAUCGGGGCAUGCGAGAUCAC ((((((((((((.((.((....)).)).))))))))))))..((((((---------------------.(---(.....)))).((((.(((((........)))))))))....)))) ( -38.60) >DroEre_CAF1 11782 96 + 1 GGUGGCCACCUUGUAGGUGUUGGCGUAGGAGGUGGCCACCGGAUGGGC---------------------CA---CAAUGGGGGCGGCGUAGCUCUUGAUGAUCGGAGCGUGCGAGAUCAG ((((((((((((.((.((....)).)).)))))))))))).(((..((---------------------(.---.......)))..(((((((((........))))).))))..))).. ( -45.60) >DroAna_CAF1 11066 99 + 1 GGUGGCCACCUUGUAGGUGUUGGCGUAGGAGGUAGCCACGGGGUGGGC---------------------CACGGCGAUGGGGGCAGCGUAGCUCUUGAUGAUCGGGGCGUGGGAGAUCAC .(((((.(((((.((.((....)).)).))))).)))))...((((.(---------------------((((.((((((((((......))))))....))))...)))))....)))) ( -37.60) >consensus GGUGGCCACCUUGUAGGUGUUGGCAUAGGAGGUGGCCACCGGGUGGGC_____________________CA___CAAUGGGCGCGGCGUAGCUCUUGAUGAUCGGGGCAUGCGAGAUCAC ((((((((((((.((.((....)).)).)))))))))))).............................................((((.(((((........)))))))))........ (-29.45 = -30.12 + 0.67)

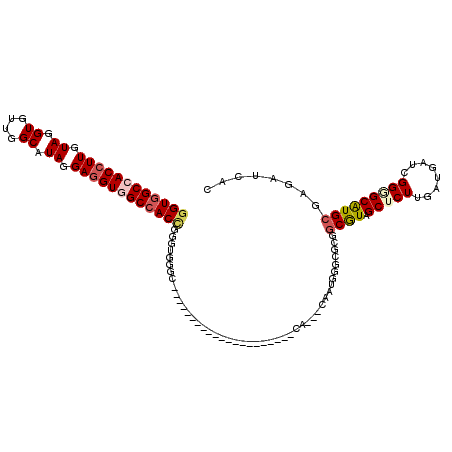
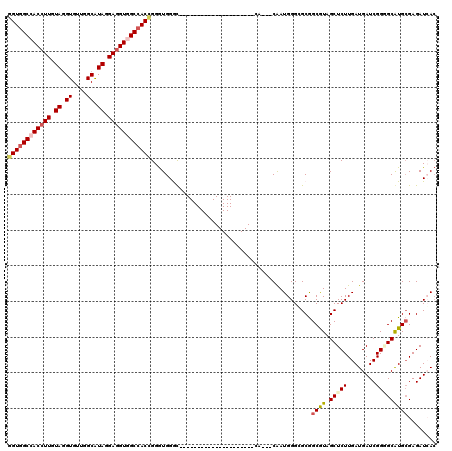
| Location | 16,260,454 – 16,260,550 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.05 |
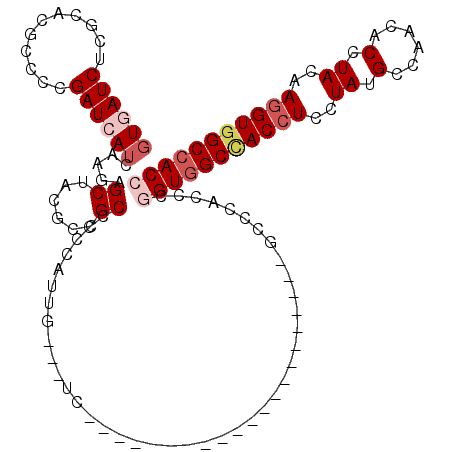
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -18.99 |
| Energy contribution | -20.22 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16260454 96 - 23771897 GUGAUCUCGCACGCCCCGAUCAUCAAGAGCUACGCCGCUCCCAUUG---UC---------------------GCCCACCCGGUGGCCACCUCCUAUGCCAACACCUACAAGGUGGCCACC (((....)))......(((.((....((((......))))....))---))---------------------).......(((((((((((..((.(......).))..))))))))))) ( -31.20) >DroVir_CAF1 11612 117 - 1 GUCAUCUCGCAUGCGCCGAUCAUCAAGAGCUAUGCCGCGCCCAUUG---UGCAUGCGCCCAUCGUGAAGACCAUCCACCCGGUGGCCACCUCCUAUGCCAACACGUACAAGGUCGCCACC (((.((.(((((((((...((.....))((......)).......)---))))))))........)).))).........((((((.((((..((((......))))..)))).)))))) ( -35.70) >DroGri_CAF1 11283 117 - 1 GUCAUCUCGCAUGCGCCGAUCAUCAAGAGCUAUGCCGCGCCCAUUG---UCCAUGCACCCAUCGUGAAGACCAUCCACCCGGUGGCCACAUCCUAUGCCAACACCUACAAGGUGGCGACC (((.....(((((.(((.........).))))))).((........---.....(((......(((..(.(((((.....)))))))))......)))...((((.....))))))))). ( -24.10) >DroSec_CAF1 11900 96 - 1 GUGAUCUCGCAUGCCCCGAUCAUCAAGAGCUACGCUGCACCCAUUG---UC---------------------GCCCACCCAGUGGCCACCUCCUAUGCCAACACCUACAAGGUGGCCACC ((((((...........))))))...(.((.(((.((....)).))---).---------------------)).).....((((((((((..((.(......).))..)))))))))). ( -25.70) >DroEre_CAF1 11782 96 - 1 CUGAUCUCGCACGCUCCGAUCAUCAAGAGCUACGCCGCCCCCAUUG---UG---------------------GCCCAUCCGGUGGCCACCUCCUACGCCAACACCUACAAGGUGGCCACC ............((((..........))))...(((((.......)---))---------------------))......(((((((((((..((.(......).))..))))))))))) ( -32.00) >DroAna_CAF1 11066 99 - 1 GUGAUCUCCCACGCCCCGAUCAUCAAGAGCUACGCUGCCCCCAUCGCCGUG---------------------GCCCACCCCGUGGCUACCUCCUACGCCAACACCUACAAGGUGGCCACC ((((((...........))))))...(.((((((..((.......))))))---------------------)).).....((((((((((..((.(......).))..)))))))))). ( -28.80) >consensus GUGAUCUCGCACGCCCCGAUCAUCAAGAGCUACGCCGCCCCCAUUG___UC_____________________GCCCACCCGGUGGCCACCUCCUAUGCCAACACCUACAAGGUGGCCACC ((((((...........)))))).....((......))..........................................(((((((((((..((.(......).))..))))))))))) (-18.99 = -20.22 + 1.22)

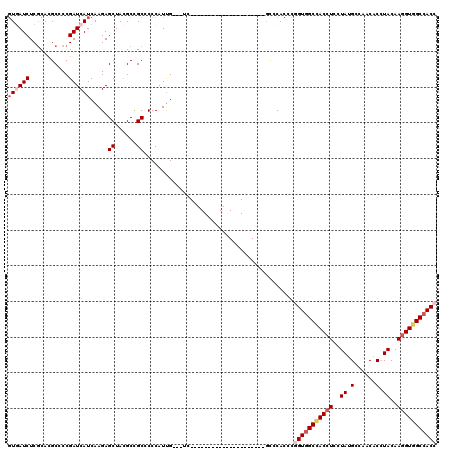
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:11 2006