
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,257,419 – 16,257,595 |
| Length | 176 |
| Max. P | 0.997912 |

| Location | 16,257,419 – 16,257,528 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.79 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.77 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16257419 109 + 23771897 CGAUAUGUUCACCUUCGCUGAACCGAAAAAGGGGGCACAGAUAGCGGGAAUUUCCACGCACGUUUUUCUUUUUGUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCA ...........((((((......)))...)))((((.......(((((.....)).)))........................((((((((....)))))))).)))). ( -30.80) >DroSec_CAF1 8859 108 + 1 CGAUAUGUUCACCUUCGCUGAACCGAAAA-AAGGGCACAGAUAGCGGGAAUUUCCACGCACGUUUUCUCUUUUCUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCA ......(((((.......)))))......-..((((.......(((((.....)).)))........................((((((((....)))))))).)))). ( -30.60) >DroSim_CAF1 7313 108 + 1 CGAUAUGUUCACCUUCGCUGAACCGAAAA-GAGGGCACAGAUAGCGGGAAUUUCCACGCACGUUUUUCUUUUCGUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCA ......(((((.......)))))......-..((((.......(((((.....)).)))........................((((((((....)))))))).)))). ( -30.60) >DroEre_CAF1 8769 96 + 1 CGAUAUGUUCACCUUCGCUGAACCGAAAA-GAGGGCACAAAU-GCGGGAAUUUCCACGCACGUUUU-----------UCCCUUCGCGCUUCAAGUGGAGCACACGCCCA ......(((((.......)))))......-..((((.....(-(((((.....)).))))......-----------.......(.(((((....))))).)..)))). ( -26.40) >consensus CGAUAUGUUCACCUUCGCUGAACCGAAAA_GAGGGCACAGAUAGCGGGAAUUUCCACGCACGUUUUUCUUUUUGUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCA ......(((((.......))))).........((((.......(((((.....)).))).........................(((((((....)))))))..)))). (-28.90 = -28.77 + -0.12)

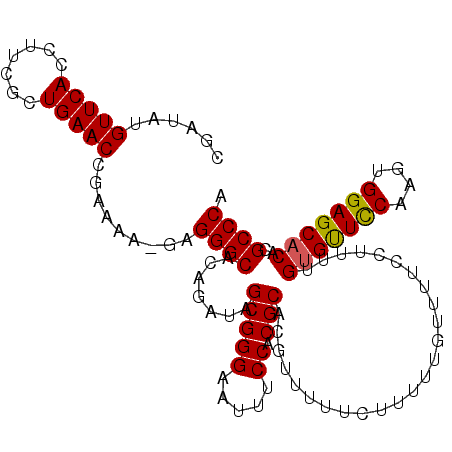
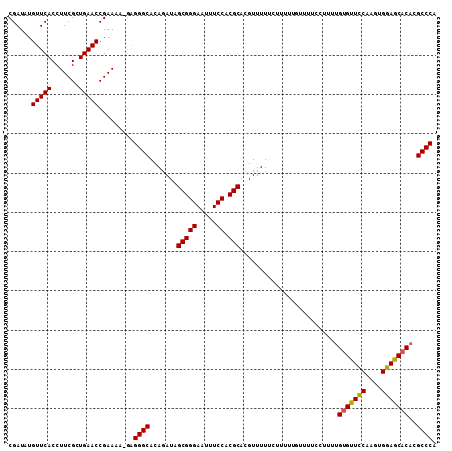
| Location | 16,257,453 – 16,257,567 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.84 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -28.21 |
| Energy contribution | -27.90 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16257453 114 + 23771897 GCACAGAUAGCGGGAAUUUCCACGCACGUUUUUCUUUUUGUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCAGCUUUUGAGCUGAUUCUACUCCUCGAGUGGUUGCUUCGC (((((((..(((((.....)).)))(((..........)))......))))))).....((((((((......(((((....)))))...((((((...)))))).)))))))) ( -32.10) >DroSec_CAF1 8892 114 + 1 GCACAGAUAGCGGGAAUUUCCACGCACGUUUUCUCUUUUCUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCAGCUCUUGAGUUGAUUCCACUCCUCGAGUGGUUGCUUCGC (((((((..(((((.....)).)))..(......)............))))))).....((((((((......(((((....)))))...((((((...)))))).)))))))) ( -32.20) >DroSim_CAF1 7346 114 + 1 GCACAGAUAGCGGGAAUUUCCACGCACGUUUUUCUUUUCGUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCAGCUCUUGAGUUGAUUCCACUCCUCGAGUGGUUGCUUCGC (((((((..(((((.....)).)))(((..........)))......))))))).....((((((((......(((((....)))))...((((((...)))))).)))))))) ( -34.10) >DroEre_CAF1 8802 102 + 1 GCACAAAU-GCGGGAAUUUCCACGCACGUUUU-----------UCCCUUCGCGCUUCAAGUGGAGCACACGCCCAGCACUUGAGCUGAUUCUGCUCCUGAAGUGGUUGCUCCGC (((....(-(((((.....)).))))......-----------........(((((((...((((((......((((......))))....)))))))))))))..)))..... ( -31.80) >consensus GCACAGAUAGCGGGAAUUUCCACGCACGUUUUUCUUUUUGUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCAGCUCUUGAGCUGAUUCCACUCCUCGAGUGGUUGCUUCGC ((.......(((((.....)).))).........................(((((((....)))))))..)).(((((....)))))...((((((...))))))......... (-28.21 = -27.90 + -0.31)

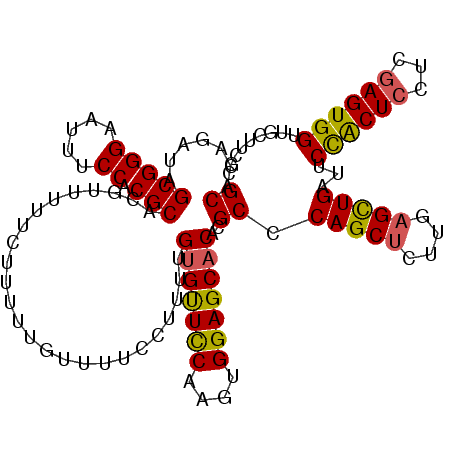
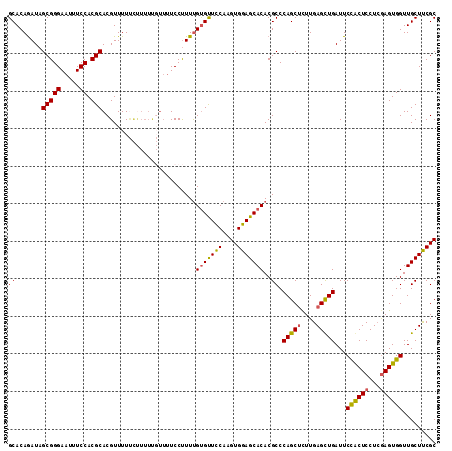
| Location | 16,257,488 – 16,257,595 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.87 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -23.19 |
| Energy contribution | -22.88 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16257488 107 + 23771897 UUUUGUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCAGCUUUUGAGCUGAUUCUACUCCUCGAGUGGUUGCUUCGCGUUUUCCCCC--AU-UUUCCA---------UACAAAUUUU .(((((.........(((((((....)))))))((((.(((((....)))))...((((((...)))))).......))))........--..-......---------.))))).... ( -27.80) >DroSec_CAF1 8927 109 + 1 UUUUCUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCAGCUCUUGAGUUGAUUCCACUCCUCGAGUGGUUGCUUCGCGUUUUCCCCCCCUU-UUUCCA---------UACAAAUUUU ...............(((((((....)))))))((((.(((((....)))))...((((((...)))))).......))))............-......---------.......... ( -26.80) >DroSim_CAF1 7381 110 + 1 UUUCGUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCAGCUCUUGAGUUGAUUCCACUCCUCGAGUGGUUGCUUCGCGUUUUCCCCCCCUUUUUUCCA---------UACAAAUUUU ...............(((((((....)))))))((((.(((((....)))))...((((((...)))))).......))))...................---------.......... ( -26.80) >DroEre_CAF1 8833 110 + 1 --------UCCCUUCGCGCUUCAAGUGGAGCACACGCCCAGCACUUGAGCUGAUUCUGCUCCUGAAGUGGUUGCUCCGCGAUUUCCCCCAUUU-UCUACACAGAUUUUCCACAAAUUUU --------.....((((((((((...((((((......((((......))))....))))))))))))((.....))))))............-......................... ( -26.40) >consensus UUUUGUUUUCCUUUUGUGUUCCAAGUGGAGCACACGCCCAGCUCUUGAGCUGAUUCCACUCCUCGAGUGGUUGCUUCGCGUUUUCCCCCCCUU_UUUCCA_________UACAAAUUUU ...............(((((((....))))))).(((.(((((....)))))...((((((...)))))).......)))....................................... (-23.19 = -22.88 + -0.31)

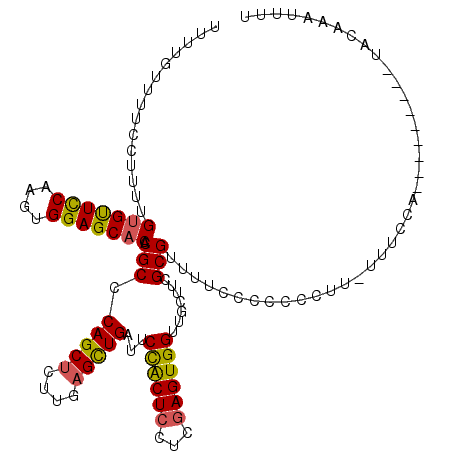

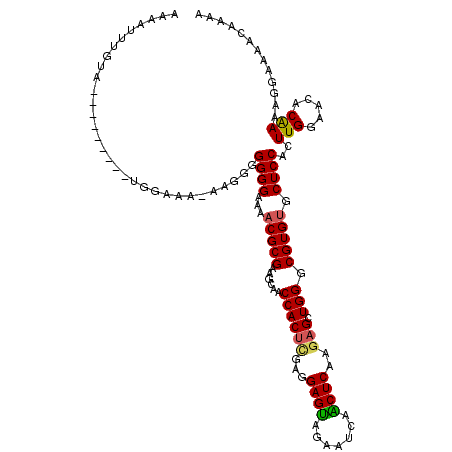
| Location | 16,257,488 – 16,257,595 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.87 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.55 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16257488 107 - 23771897 AAAAUUUGUA---------UGGAAA-AU--GGGGGAAAACGCGAAGCAACCACUCGAGGAGUAGAAUCAGCUCAAAAGCUGGGCGUGUGCUCCACUUGGAACACAAAAGGAAAACAAAA ....(((((.---------......-..--.((((...((((.........((((...))))....((((((....))))))))))...))))..(((.....))).......))))). ( -23.90) >DroSec_CAF1 8927 109 - 1 AAAAUUUGUA---------UGGAAA-AAGGGGGGGAAAACGCGAAGCAACCACUCGAGGAGUGGAAUCAACUCAAGAGCUGGGCGUGUGCUCCACUUGGAACACAAAAGGAAAAGAAAA ....(((((.---------......-(((..((((...((((..(((..((((((...))))))..((.......)))))..))))...)))).))).....)))))............ ( -28.40) >DroSim_CAF1 7381 110 - 1 AAAAUUUGUA---------UGGAAAAAAGGGGGGGAAAACGCGAAGCAACCACUCGAGGAGUGGAAUCAACUCAAGAGCUGGGCGUGUGCUCCACUUGGAACACAAAAGGAAAACGAAA ....(((((.---------.......(((..((((...((((..(((..((((((...))))))..((.......)))))..))))...)))).))).....)))))............ ( -27.82) >DroEre_CAF1 8833 110 - 1 AAAAUUUGUGGAAAAUCUGUGUAGA-AAAUGGGGGAAAUCGCGGAGCAACCACUUCAGGAGCAGAAUCAGCUCAAGUGCUGGGCGUGUGCUCCACUUGAAGCGCGAAGGGA-------- ....(((((((....((((((....-.............))))))....)).(((((((((((.(.(((((......)))))...).))))))...))))).)))))....-------- ( -32.13) >consensus AAAAUUUGUA_________UGGAAA_AAGGGGGGGAAAACGCGAAGCAACCACUCGAGGAGUAGAAUCAACUCAAGAGCUGGGCGUGUGCUCCACUUGGAACACAAAAGGAAAACAAAA ...............................((((...(((((......((((((...((((.......))))..))).))).))))).))))..(((.....)))............. (-20.55 = -20.55 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:08 2006