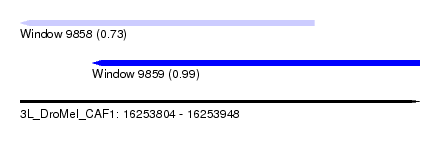
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,253,804 – 16,253,948 |
| Length | 144 |
| Max. P | 0.989568 |

| Location | 16,253,804 – 16,253,910 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 87.05 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -21.69 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16253804 106 - 23771897 AAACAAAGGCAGCUACAAAUGCCAAAAAAGCUUCGUCUUGGCCUGCACUCAAAGUGUUGGCCCAAAGCCGUUCACCAAGAGCCAAAAGAAGACCUGAACAGAG----------UGG .......((((........)))).......(((((..((((((.((((.....)))).)).))))..).((((.....)))).....))))............----------... ( -26.00) >DroSec_CAF1 5299 105 - 1 AAACAAAGGCAGCCACAAAUGCCAAAAAAGCUUCGUCUUGGCCUGCACUCA-AGUGUUGGCCCAAAGCCGUUCACAAAGAGCCAAAAGAAGACCUGAACAGAG----------UGG .......((((........)))).......(((((..((((((.((((...-.)))).)).))))..).((((.....)))).....))))............----------... ( -26.00) >DroSim_CAF1 3708 105 - 1 AAACAAAGGCAGCCACAAAUGCCAAAAAAGCUUCGUCUUGGCCUGCACUCA-AGUGUUGGCCCAAAGCCGUUCACAAAGAGCCAAAAGAAGACCUGAACAGAG----------UGG .......((((........)))).......(((((..((((((.((((...-.)))).)).))))..).((((.....)))).....))))............----------... ( -26.00) >DroEre_CAF1 5257 102 - 1 AAACAAAGGCAGCA--AAAUGCCAA-AAAGCUUCGUCUUGGCCUGCACUCA-AGUGUUGGCUCAAAGCCGUUCACAAAGAGCCAAAAGAAGACCUGAAAAGAG----------UGG .......((((...--...))))..-....(((((..((((((.((((...-.)))).))).)))..).((((.....)))).....))))((((....)).)----------).. ( -27.90) >DroYak_CAF1 7111 102 - 1 AAACAAAGGCAGCU--AAAUGCCAA-AAAGCUUCGUCUUGGCCUGCACUCA-AGUGUUGGCUCAAAGCCGUUCACAAAGAGCCAAAAGAAGACCUAAAAAGAG----------UGG .......((((...--...))))..-....(((((..((((((.((((...-.)))).))).)))..).((((.....)))).....))))............----------... ( -26.20) >DroAna_CAF1 3918 106 - 1 AAACAAAGGCAACA--AAAUGCCAA-AAAGCUUCGUCUUGGCCUGCACUCA-AGCGUUGGCUUAAA--CAUUCAC----AGCCAAAAAGUAGCAUCAAAAGAGACAGAAAUAGGGG .......((((...--...))))..-.....((((((((....((((((..-....((((((....--.......----))))))..))).)))......))))).)))....... ( -20.90) >consensus AAACAAAGGCAGCA__AAAUGCCAA_AAAGCUUCGUCUUGGCCUGCACUCA_AGUGUUGGCCCAAAGCCGUUCACAAAGAGCCAAAAGAAGACCUGAAAAGAG__________UGG .......((((........))))...........(((((((((.((((.....)))).)))).......((((.....))))......)))))....................... (-21.69 = -22.00 + 0.31)
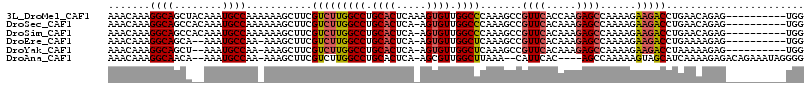
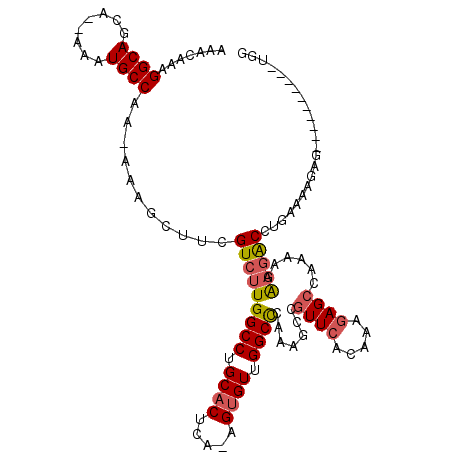
| Location | 16,253,830 – 16,253,948 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.69 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -23.74 |
| Energy contribution | -23.71 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16253830 118 - 23771897 ACCGCUUUGUUAAGCCACCAG--AAAUAAACAAAGUAAAUAAACAAAGGCAGCUACAAAUGCCAAAAAAGCUUCGUCUUGGCCUGCACUCAAAGUGUUGGCCCAAAGCCGUUCACCAAGA ((.((((((..((((......--...........((......))...((((........))))......))))......((((.((((.....)))).)))))))))).))......... ( -27.70) >DroSec_CAF1 5325 117 - 1 ACCGCUUUGUUAAGCCACCAG--AAAUAAACAAAGUAAAUAAACAAAGGCAGCCACAAAUGCCAAAAAAGCUUCGUCUUGGCCUGCACUCA-AGUGUUGGCCCAAAGCCGUUCACAAAGA ((.((((((..((((......--...........((......))...((((........))))......))))......((((.((((...-.)))).)))))))))).))......... ( -27.70) >DroSim_CAF1 3734 117 - 1 ACCGCUUUGUUAAGCCACCAG--AAAUAAACAAAGUAAAUAAACAAAGGCAGCCACAAAUGCCAAAAAAGCUUCGUCUUGGCCUGCACUCA-AGUGUUGGCCCAAAGCCGUUCACAAAGA ((.((((((..((((......--...........((......))...((((........))))......))))......((((.((((...-.)))).)))))))))).))......... ( -27.70) >DroEre_CAF1 5283 114 - 1 ACCGCUUUGUUAAGCCAUCAG--AAACAAACAAAGUAAACAAACAAAGGCAGCA--AAAUGCCAA-AAAGCUUCGUCUUGGCCUGCACUCA-AGUGUUGGCUCAAAGCCGUUCACAAAGA ....((((((.((((......--...........((......))...((((...--...))))..-...)))).(..((((((.((((...-.)))).))).)))..).....)))))). ( -26.30) >DroYak_CAF1 7137 114 - 1 ACCGCUUUGUUAAGCCACCAG--AAAUAAACAAAGUAAAUAAACAAAGGCAGCU--AAAUGCCAA-AAAGCUUCGUCUUGGCCUGCACUCA-AGUGUUGGCUCAAAGCCGUUCACAAAGA ....((((((.((((......--...........((......))...((((...--...))))..-...)))).(..((((((.((((...-.)))).))).)))..).....)))))). ( -26.30) >DroAna_CAF1 3954 110 - 1 ACAGCUUUGUUGAGUCAACUGACAAACAAACAAAGCAAAUAAACAAAGGCAACA--AAAUGCCAA-AAAGCUUCGUCUUGGCCUGCACUCA-AGCGUUGGCUUAAA--CAUUCAC----A ...((((((((..(((....))).....))))))))...........((((...--...))))..-.(((((.((.(((((.......)))-))))..)))))...--.......----. ( -27.40) >consensus ACCGCUUUGUUAAGCCACCAG__AAAUAAACAAAGUAAAUAAACAAAGGCAGCA__AAAUGCCAA_AAAGCUUCGUCUUGGCCUGCACUCA_AGUGUUGGCCCAAAGCCGUUCACAAAGA ...((((((((.................))))))))...........((((........))))......((((.(....((((.((((.....)))).))))).))))............ (-23.74 = -23.71 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:02 2006