
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,225,933 – 16,226,133 |
| Length | 200 |
| Max. P | 0.896847 |

| Location | 16,225,933 – 16,226,053 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -40.51 |
| Consensus MFE | -31.15 |
| Energy contribution | -31.78 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16225933 120 + 23771897 UUAUUAACAAUGCUAAGUAUCAAUGGCCAUAGAGCAAUCUACGCAUUUGCGUAUGUUCCAUGGGUGGAUGCCAUGUGACGGCCUCGGGUCACGUUCCGGAGCGAUUCCUUUCGGGAUCGA ...........(((..........((((.((((....)))).(((((..(.((((...)))).)..)))))........))))(((((......))))))))((((((....)))))).. ( -39.50) >DroSec_CAF1 44729 120 + 1 UUAUUAUCCCUACUAAAUAUCAAUGGCCAUAGAACAAUCUAUGCAUUCGCGUAUGUUCCAUGGGUGGAUGCCAUGUGACGGCCUCGGGUCACGUUCCGGAGCGAUUCCUUUCGGGAUCGA ......(((...............(((((((((....)))))((((((((.((((...)))).))))))))........))))..(((......)))))).(((((((....))))))). ( -38.70) >DroSim_CAF1 46441 120 + 1 UUAUUAUCCCUGCUAAGUAUCAAUGGCCAUAGAGCAAUCUACGCAUUCGCGUAUGUUCUAUGGGUGGAUGCCAUGUGACGGCCUCGGGUCACGUUCCGGAGCGAUUCCUUUCGGGAUCGA ......(((.......(((((...(.((((((((((...(((((....))))))))))))))).).))))).(((((((........)))))))...))).(((((((....))))))). ( -44.70) >DroEre_CAF1 44514 120 + 1 UAACUAACCCUGCUGAGUAUCGAUGGCCAUCCAGCAAUCUACGCACUUGGGUAUCUUCCAUGGCUGGAUGCCAUGUGACAACCUCGGGUCACGUUCCGAAGCGAUUCCCUUCGGGAUCGA ......((((....(((..((((((((.(((((((............((((.....))))..)))))))))))).)))....)))))))..((((((((((.(....))))))))).)). ( -39.14) >consensus UUAUUAACCCUGCUAAGUAUCAAUGGCCAUAGAGCAAUCUACGCAUUCGCGUAUGUUCCAUGGGUGGAUGCCAUGUGACGGCCUCGGGUCACGUUCCGGAGCGAUUCCUUUCGGGAUCGA ................(((((...(.((((.(((((...(((((....)))))))))).)))).).))))).(((((((........))))))).......(((((((....))))))). (-31.15 = -31.78 + 0.63)



| Location | 16,225,933 – 16,226,053 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -28.26 |
| Energy contribution | -28.70 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16225933 120 - 23771897 UCGAUCCCGAAAGGAAUCGCUCCGGAACGUGACCCGAGGCCGUCACAUGGCAUCCACCCAUGGAACAUACGCAAAUGCGUAGAUUGCUCUAUGGCCAUUGAUACUUAGCAUUGUUAAUAA ..(((.((....)).)))(((.(((........))).((((((..(((((.......)))))..)).(((((....)))))...........))))..........)))........... ( -36.00) >DroSec_CAF1 44729 120 - 1 UCGAUCCCGAAAGGAAUCGCUCCGGAACGUGACCCGAGGCCGUCACAUGGCAUCCACCCAUGGAACAUACGCGAAUGCAUAGAUUGUUCUAUGGCCAUUGAUAUUUAGUAGGGAUAAUAA ...(((((..........((..(((........)))..)).((((.(((((.((((....)))).............((((((....)))))))))))))))........)))))..... ( -35.10) >DroSim_CAF1 46441 120 - 1 UCGAUCCCGAAAGGAAUCGCUCCGGAACGUGACCCGAGGCCGUCACAUGGCAUCCACCCAUAGAACAUACGCGAAUGCGUAGAUUGCUCUAUGGCCAUUGAUACUUAGCAGGGAUAAUAA ...(((((....(((...((..(((........)))..)).(((....))).)))..(((((((.(((((((....)))))...)).)))))))................)))))..... ( -36.10) >DroEre_CAF1 44514 120 - 1 UCGAUCCCGAAGGGAAUCGCUUCGGAACGUGACCCGAGGUUGUCACAUGGCAUCCAGCCAUGGAAGAUACCCAAGUGCGUAGAUUGCUGGAUGGCCAUCGAUACUCAGCAGGGUUAGUUA ......((((((.(...).))))))(((.((((((..((.((((.((((((.....))))))...)))).))...(((..((((((.(((....))).)))).))..)))))))))))). ( -42.90) >consensus UCGAUCCCGAAAGGAAUCGCUCCGGAACGUGACCCGAGGCCGUCACAUGGCAUCCACCCAUGGAACAUACGCAAAUGCGUAGAUUGCUCUAUGGCCAUUGAUACUUAGCAGGGAUAAUAA ..(((.((....)).)))(((.(((........))).((((((..(((((.......)))))..)).(((((....)))))...........))))..........)))........... (-28.26 = -28.70 + 0.44)

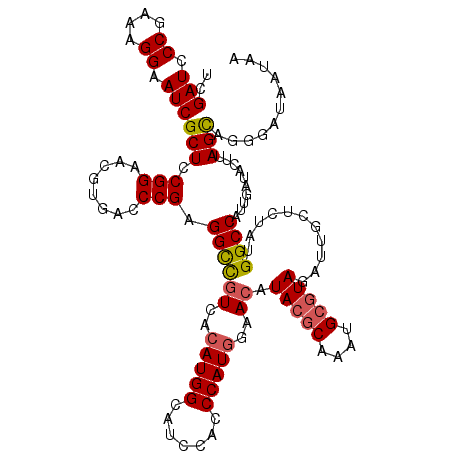

| Location | 16,226,013 – 16,226,133 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -49.42 |
| Consensus MFE | -45.85 |
| Energy contribution | -45.23 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16226013 120 + 23771897 GCCUCGGGUCACGUUCCGGAGCGAUUCCUUUCGGGAUCGACGAGCCACUCGCCAAAGGGCAAAAAUCGGGGGAACAGCCUGUGUGAGUCACAUUGCACCUUGCGGUGGACUCUACGGCUU (((.(((((...(((((((.((((((((....))))))).)...))....(((....)))..........))))).))))).(((((((.((((((.....))))))))))).))))).. ( -49.80) >DroSec_CAF1 44809 120 + 1 GCCUCGGGUCACGUUCCGGAGCGAUUCCUUUCGGGAUCGACGAGCCGUUCGCCAAAGGGCACAAAUCGAGGGAGCAGCCUGUGGGAGUCACAUUGCACCUUGCGGUGGACUCUACGGCUU (((.(((((...(((((((.((((((((....))))))).)...))....(((....)))..........))))).)))))..((((((.((((((.....))))))))))))..))).. ( -50.30) >DroSim_CAF1 46521 120 + 1 GCCUCGGGUCACGUUCCGGAGCGAUUCCUUUCGGGAUCGACGAGCCAUUCGCCAAAGGGCAAAAAUCGAGGGAGCAGCCUGUGGGAGUCACAUUGCACCUUGCGGUGGACUCUACGGCUU (((.(((((...(((((((.((((((((....))))))).)...))....(((....)))..........))))).)))))..((((((.((((((.....))))))))))))..))).. ( -50.30) >DroEre_CAF1 44594 119 + 1 ACCUCGGGUCACGUUCCGAAGCGAUUCCCUUCGGGAUCGACAUGCCAUUUGCCAAAGGGCAAAAAUCG-GGGAGCAGCUUGUGGGAGUCACAUUGCACCUUGCGGUGGAUUCUACGGCUU .(((((((((..(((((((((.(....)))))))))).)))......((((((....))))))..)))-)))...((((.((((.((((.((((((.....)))))))))))))))))). ( -47.30) >consensus GCCUCGGGUCACGUUCCGGAGCGAUUCCUUUCGGGAUCGACGAGCCAUUCGCCAAAGGGCAAAAAUCGAGGGAGCAGCCUGUGGGAGUCACAUUGCACCUUGCGGUGGACUCUACGGCUU .(((((((((..(((((((((.(....)))))))))).))).........(((....))).....))))))....((((.(((.(((((.((((((.....)))))))))))))))))). (-45.85 = -45.23 + -0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:54 2006