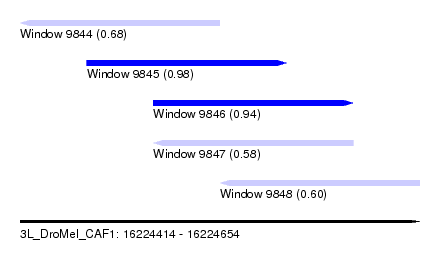
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,224,414 – 16,224,654 |
| Length | 240 |
| Max. P | 0.975755 |

| Location | 16,224,414 – 16,224,534 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -39.09 |
| Consensus MFE | -33.17 |
| Energy contribution | -34.48 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16224414 120 - 23771897 GCUACCAGCAGCCUUACCUGGGCCCCCAGCCAGAAUCCUACUACUACGGAGGACGGUACCCGGCUGGUGGCUACUACAACAACUACCACAACAGCCCGGGUCCGUACGGCUACCCCUAUC ((.....)).((((.....))))....((((.....(((.((.....)))))((((.(((((((((((((...............))))..))).))))))))))..))))......... ( -39.26) >DroSec_CAF1 43200 120 - 1 GCUACCAGCAGCCUUACCUGGGCCCCCAGCCAGGAUCCUACUACUACGGAGGACCGUACCCGGCUGGUGGCUACUACAACAACUACCACAACAGCCCGGGUCCGUACGGCUACCCCUAUC ((.....))((((.(((((((....))))...((((((......(((((....)))))...(((((((((...............))))..))))).))))))))).))))......... ( -41.36) >DroSim_CAF1 44851 120 - 1 GCUACCAGCAGCCUUACCUGGGCCCCCAGCCAGGAUCCUACUACUACGGAGGACCGUACCCGGCUGGUGGCUACUACAACAACUACCACAACAGCCCGGGUCCGUACGGCUACCCCUAUC ((.....))((((.(((((((....))))...((((((......(((((....)))))...(((((((((...............))))..))))).))))))))).))))......... ( -41.36) >DroEre_CAF1 43009 120 - 1 GCUACCAGCAGCCUUACCUGGGCCCCCAGCCAGGACCCUACUACUACGGAGAAGCGUACCCGGCAGGUGGCUACUACAACAACUACCACAACAGCCCGGGACCGUACGGAUACCCCUAUC ((.....)).......(((((((.....(((.((((.((.((.(....))).)).)).)).))).(((((............)))))......))))))).((....))........... ( -34.40) >consensus GCUACCAGCAGCCUUACCUGGGCCCCCAGCCAGGAUCCUACUACUACGGAGGACCGUACCCGGCUGGUGGCUACUACAACAACUACCACAACAGCCCGGGUCCGUACGGCUACCCCUAUC ((.....))((((.(((((((....))))...((((((......(((((....)))))...(((((((((...............))))..))))).))))))))).))))......... (-33.17 = -34.48 + 1.31)

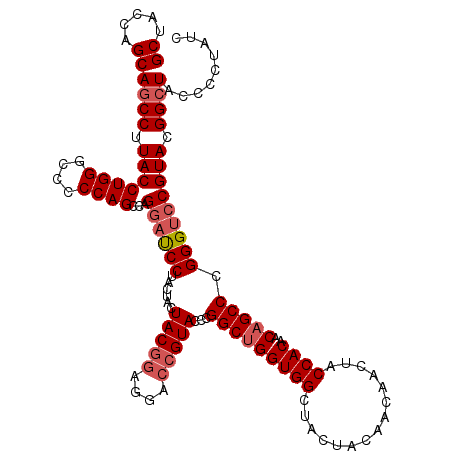
| Location | 16,224,454 – 16,224,574 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -55.83 |
| Consensus MFE | -55.34 |
| Energy contribution | -54.90 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16224454 120 + 23771897 GUUGUAGUAGCCACCAGCCGGGUACCGUCCUCCGUAGUAGUAGGAUUCUGGCUGGGGGCCCAGGUAAGGCUGCUGGUAGCUGACACCGAAGGAGACCAGCGGCUUGGCCUUGAUGCAGGU .(((((...(((.(((((((((....(((((.(......).)))))))))))))).)))(.((((.(((((((((((..((.........))..))))))))))).)))).).))))).. ( -55.50) >DroSec_CAF1 43240 120 + 1 GUUGUAGUAGCCACCAGCCGGGUACGGUCCUCCGUAGUAGUAGGAUCCUGGCUGGGGGCCCAGGUAAGGCUGCUGGUAGCUGACGCCGAAGGAGACCAGUGGCUUGGCCUUGAUGCAGGU .(((((...(((.(((((((((....(((((.(......).)))))))))))))).)))(.((((.((((..(((((..((.........))..)))))..)))).)))).).))))).. ( -55.30) >DroSim_CAF1 44891 120 + 1 GUUGUAGUAGCCACCAGCCGGGUACGGUCCUCCGUAGUAGUAGGAUCCUGGCUGGGGGCCCAGGUAAGGCUGCUGGUAGCUGACGCCGAAGGAGACCAGCGGCUUGGCCUUGAUGCAGGU .(((((...(((.(((((((((....(((((.(......).)))))))))))))).)))(.((((.(((((((((((..((.........))..))))))))))).)))).).))))).. ( -57.80) >DroEre_CAF1 43049 120 + 1 GUUGUAGUAGCCACCUGCCGGGUACGCUUCUCCGUAGUAGUAGGGUCCUGGCUGGGGGCCCAGGUAAGGCUGCUGGUAGCCGACGCCGAAGGAGACCAGCGGCUUGGCCUUGAUGCAGGU .(((((...(((.(((((((((.((.((.((.......)).)).)))))))).))))))(.((((.(((((((((((..((.........))..))))))))))).)))).).))))).. ( -54.70) >consensus GUUGUAGUAGCCACCAGCCGGGUACGGUCCUCCGUAGUAGUAGGAUCCUGGCUGGGGGCCCAGGUAAGGCUGCUGGUAGCUGACGCCGAAGGAGACCAGCGGCUUGGCCUUGAUGCAGGU .(((((...(((.(((((((((....(((((.(......).)))))))))))))).)))(.((((.(((((((((((..((.........))..))))))))))).)))).).))))).. (-55.34 = -54.90 + -0.44)

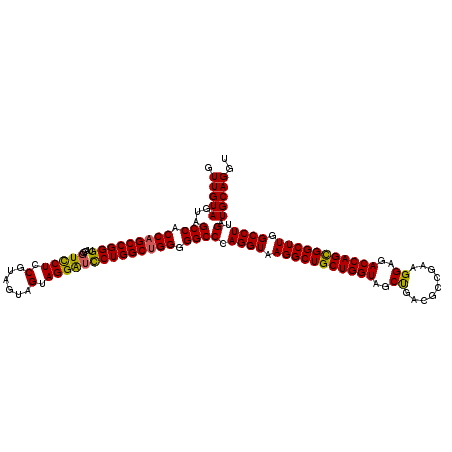

| Location | 16,224,494 – 16,224,614 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -52.33 |
| Consensus MFE | -48.74 |
| Energy contribution | -48.42 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16224494 120 + 23771897 UAGGAUUCUGGCUGGGGGCCCAGGUAAGGCUGCUGGUAGCUGACACCGAAGGAGACCAGCGGCUUGGCCUUGAUGCAGGUGGUGGCCACCACCAGGAGCACUGCUAACUGCAACGGUAGA (((..((((((.(((.((((.((((.(((((((((((..((.........))..))))))))))).))))....((.....)))))).)))))))))...)))....((((....)))). ( -49.50) >DroSec_CAF1 43280 120 + 1 UAGGAUCCUGGCUGGGGGCCCAGGUAAGGCUGCUGGUAGCUGACGCCGAAGGAGACCAGUGGCUUGGCCUUGAUGCAGGUGGUGGCCACCACCAGGAGCACUGCUAACUGCAAGGUAUGA .....((((((.(((.((((.((((.((((..(((((..((.........))..)))))..)))).))))....((.....)))))).)))))))))((..(((.....)))..)).... ( -49.20) >DroSim_CAF1 44931 120 + 1 UAGGAUCCUGGCUGGGGGCCCAGGUAAGGCUGCUGGUAGCUGACGCCGAAGGAGACCAGCGGCUUGGCCUUGAUGCAGGUGGUGGCCACCACCAGGAGCACUGCUAACUGCAAGGUAAGA .....((((((.(((.((((.((((.(((((((((((..((.........))..))))))))))).))))....((.....)))))).)))))))))((..(((.....)))..)).... ( -51.70) >DroEre_CAF1 43089 120 + 1 UAGGGUCCUGGCUGGGGGCCCAGGUAAGGCUGCUGGUAGCCGACGCCGAAGGAGACCAGCGGCUUGGCCUUGAUGCAGGUGGUGGCCACCAGCAGGAGCACUGCUAACUGCAAUAGGAGG ..(((((((.....)))))))((((.(((((((((((..((.........))..))))))))))).))))...(((((((((...)))))(((((.....)))))...))))........ ( -58.90) >consensus UAGGAUCCUGGCUGGGGGCCCAGGUAAGGCUGCUGGUAGCUGACGCCGAAGGAGACCAGCGGCUUGGCCUUGAUGCAGGUGGUGGCCACCACCAGGAGCACUGCUAACUGCAAGGGAAGA ...(.((((((.(((.((((.((((.(((((((((((..((.........))..))))))))))).))))....((.....)))))).))))))))).)..(((.....)))........ (-48.74 = -48.42 + -0.31)

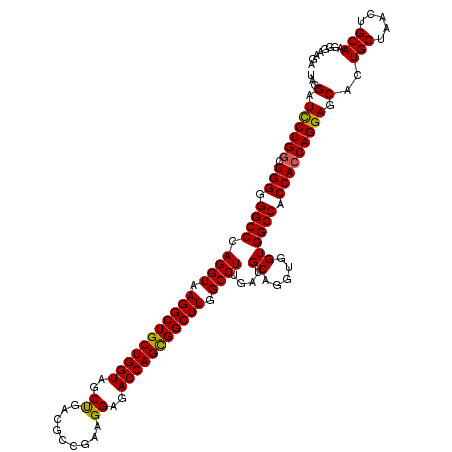
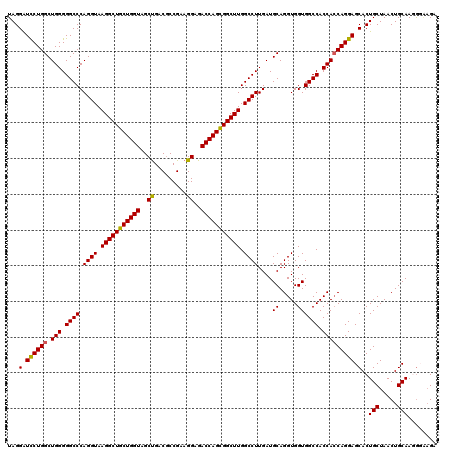
| Location | 16,224,494 – 16,224,614 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -42.24 |
| Consensus MFE | -37.60 |
| Energy contribution | -38.41 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16224494 120 - 23771897 UCUACCGUUGCAGUUAGCAGUGCUCCUGGUGGUGGCCACCACCUGCAUCAAGGCCAAGCCGCUGGUCUCCUUCGGUGUCAGCUACCAGCAGCCUUACCUGGGCCCCCAGCCAGAAUCCUA (((...(((((.(.((((.((((....((((((....)))))).))))...(((...((((..((...))..))))))).)))).).))))).....((((....))))..)))...... ( -39.10) >DroSec_CAF1 43280 120 - 1 UCAUACCUUGCAGUUAGCAGUGCUCCUGGUGGUGGCCACCACCUGCAUCAAGGCCAAGCCACUGGUCUCCUUCGGCGUCAGCUACCAGCAGCCUUACCUGGGCCCCCAGCCAGGAUCCUA .......((((.....))))...(((((((((.((((.....((((....((((((......)))))).....(((....)))....)))).........)))).))).))))))..... ( -39.14) >DroSim_CAF1 44931 120 - 1 UCUUACCUUGCAGUUAGCAGUGCUCCUGGUGGUGGCCACCACCUGCAUCAAGGCCAAGCCGCUGGUCUCCUUCGGCGUCAGCUACCAGCAGCCUUACCUGGGCCCCCAGCCAGGAUCCUA .......((((.....))))...(((((((((.((((....(((......)))....((.((((((.......(((....))))))))).))........)))).))).))))))..... ( -41.91) >DroEre_CAF1 43089 120 - 1 CCUCCUAUUGCAGUUAGCAGUGCUCCUGCUGGUGGCCACCACCUGCAUCAAGGCCAAGCCGCUGGUCUCCUUCGGCGUCGGCUACCAGCAGCCUUACCUGGGCCCCCAGCCAGGACCCUA .....((((((.....))))))...((((((((((((....(((......)))....((((..((...))..))))...))))))))))))(((...((((....))))..)))...... ( -48.80) >consensus UCUUACCUUGCAGUUAGCAGUGCUCCUGGUGGUGGCCACCACCUGCAUCAAGGCCAAGCCGCUGGUCUCCUUCGGCGUCAGCUACCAGCAGCCUUACCUGGGCCCCCAGCCAGGAUCCUA .....((((((.....)))))).(((((((((.((((....(((......)))....((.((((((.......(((....))))))))).))........)))).))).))))))..... (-37.60 = -38.41 + 0.81)

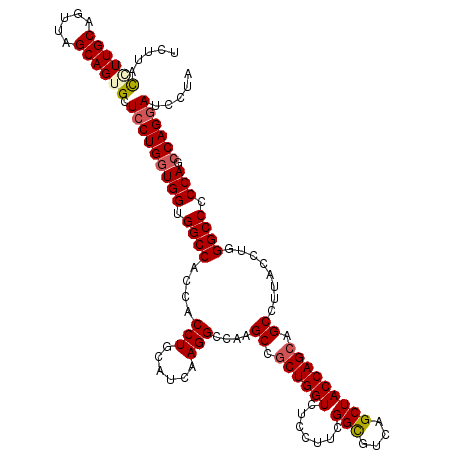
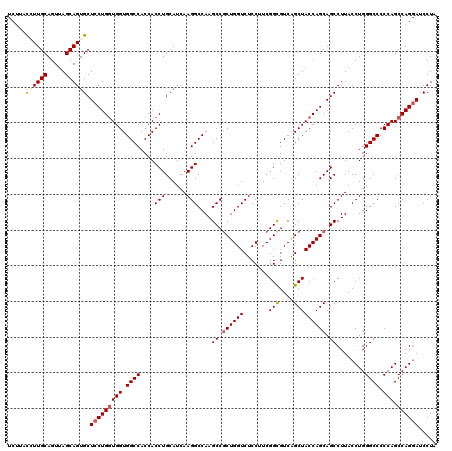
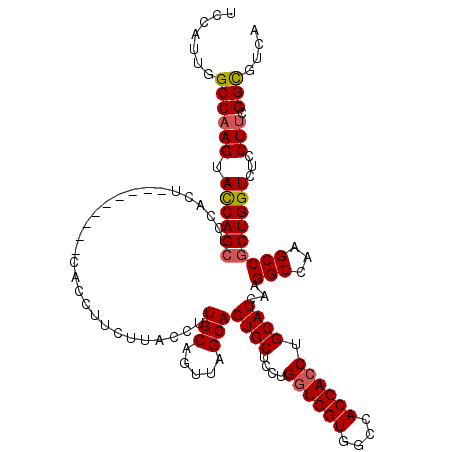
| Location | 16,224,534 – 16,224,654 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.16 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -29.35 |
| Energy contribution | -29.73 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16224534 120 - 23771897 UCCAUUGGCCAAGUAUCAGCUCCACUUAACUUCUCCACCUUCUACCGUUGCAGUUAGCAGUGCUCCUGGUGGUGGCCACCACCUGCAUCAAGGCCAAGCCGCUGGUCUCCUUCGGUGUCA ....((((((........(((..(((((((................)))).))).))).((((....((((((....)))))).))))...))))))(.((((((......)))))).). ( -34.59) >DroSec_CAF1 43320 111 - 1 UCCAUUGGCCAAGUACCAGCUCCACU---------CACCUUCAUACCUUGCAGUUAGCAGUGCUCCUGGUGGUGGCCACCACCUGCAUCAAGGCCAAGCCACUGGUCUCCUUCGGCGUCA ....((((((........(((..(((---------((...........)).))).))).((((....((((((....)))))).))))...))))))(((...((...))...))).... ( -31.70) >DroSim_CAF1 44971 111 - 1 UCCAUUGGCCAAGUACCAGCUCCACU---------CACCUUCUUACCUUGCAGUUAGCAGUGCUCCUGGUGGUGGCCACCACCUGCAUCAAGGCCAAGCCGCUGGUCUCCUUCGGCGUCA ....((((((........(((..(((---------((...........)).))).))).((((....((((((....)))))).))))...))))))((((..((...))..)))).... ( -35.10) >DroEre_CAF1 43129 114 - 1 UCCAUCAGCC-AGUACCAGCUCCUCUCAG-----CCACCUCCUCCUAUUGCAGUUAGCAGUGCUCCUGCUGGUGGCCACCACCUGCAUCAAGGCCAAGCCGCUGGUCUCCUUCGGCGUCG .......(((-((.((((((....((..(-----((.........((((((.....))))))....(((.(((((...))))).)))....)))..))..)))))))).....))).... ( -34.50) >consensus UCCAUUGGCCAAGUACCAGCUCCACU_________CACCUUCUUACCUUGCAGUUAGCAGUGCUCCUGGUGGUGGCCACCACCUGCAUCAAGGCCAAGCCGCUGGUCUCCUUCGGCGUCA .......((((((.((((((............................(((.....)))((((....((((((....)))))).))))...(((...)))))))))...))).))).... (-29.35 = -29.73 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:51 2006