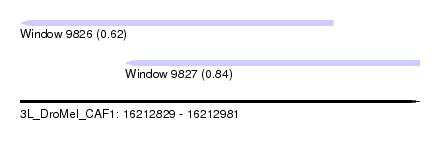
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,212,829 – 16,212,981 |
| Length | 152 |
| Max. P | 0.838693 |

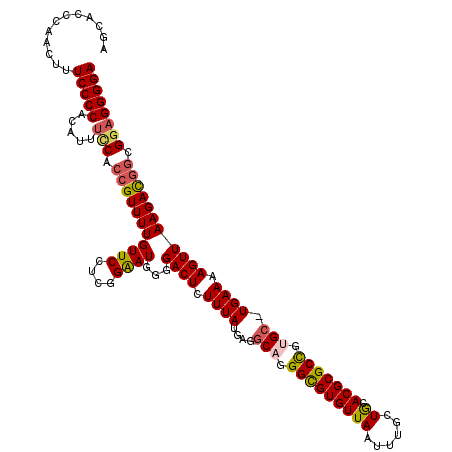
| Location | 16,212,829 – 16,212,948 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.89 |
| Mean single sequence MFE | -41.90 |
| Consensus MFE | -31.80 |
| Energy contribution | -31.00 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16212829 119 - 23771897 GGGAUGGGGACUCUUUAUGAGUCAGGGCGUGUUAAUUUGCUGCACGCGCCGUGC-UGAAAAGUUAAGACGGCGGAGGGGAUCAAGGAUGCGGUCAACUUGUUGCCAGUCCACCAAAUUGA ....(((.(((((.....))))).((((((.(((((((...(((((...)))))-....))))))).))(((((((..((((........))))..)))..)))).)))).)))...... ( -42.80) >DroSec_CAF1 31584 119 - 1 GGAAUGGGGACUCUUUAUGGGGCAGGGCGUGUUAAUUUGCUGCACGCGCCGUGC-UGAAAAGUUAAGAUGGCGGAGGGGAUCAAGGAUGCGGUCAACUUGUUGCCAGUCCACCAAAUUGA ....(((((((((((.((..((((.(((((((((......)).))))))).)))-).....)).))))((((((((..((((........))))..)))..))))))))).)))...... ( -42.00) >DroSim_CAF1 33427 119 - 1 GGAAUGGGGACUCUUUAUGGGGCAGGGCGUGUUAAUUUGCUGCACGCGCCGUGC-UGAAAAGUUAAGACGGCGGAGGGGAUCAAGGAUGCGGUCAACUUGUUGCCAGUCCACCAAAUUUA ....((((((((((....))(((((.((((((.........))))))(((((((-(....)))....))))).(((..((((........))))..))).)))))))))).)))...... ( -42.20) >DroEre_CAF1 31068 120 - 1 GGGAUGGGGACUCUUUAUGAGGCAGGGCGUGUUAAUUUGUUGCACGCGCCGGGCAUGAAAAGUUAAGACGGCGGCGGGGAUCAAGGAGGUGGUCAACUUGUUGCCACUCCACCGCUUUAA .((.((((((((.((((((.(((...((((((.........)))))))))...)))))).)))......((((((((((((((......)))))..))))))))).)))))))....... ( -45.00) >DroYak_CAF1 68127 120 - 1 GGAAUGGGGACUCUUUAUGAGGCAGGGUGUGUUAAUUUGGUACACGCGCUGGGCAUGAAAAGUUAAGAUGGGGGAGGGGAUCAAGGAUCCGGUCAACUUGUUGCUACUCCACCGCUUUAA ........((((.((((((...(((.((((((.........)))))).)))..)))))).))))(((.(((.((((.(((((...)))))((.(((....))))).)))).))))))... ( -37.50) >consensus GGAAUGGGGACUCUUUAUGAGGCAGGGCGUGUUAAUUUGCUGCACGCGCCGUGC_UGAAAAGUUAAGACGGCGGAGGGGAUCAAGGAUGCGGUCAACUUGUUGCCAGUCCACCAAAUUAA (((.(((.(((..........(((.(((((((((......)).))))))).))).....(((((..(((.(((..............))).))))))))))).))).))).......... (-31.80 = -31.00 + -0.80)

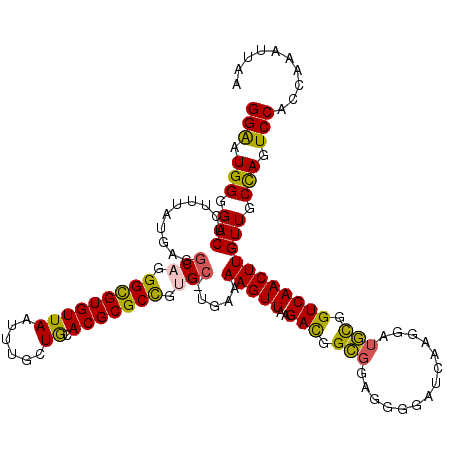
| Location | 16,212,869 – 16,212,981 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.05 |
| Mean single sequence MFE | -44.76 |
| Consensus MFE | -33.94 |
| Energy contribution | -34.26 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16212869 112 - 23771897 AG-------CUUUCCCCACAUUUCCACCGUUUUGUUCCUCGGGAUGGGGACUCUUUAUGAGUCAGGGCGUGUUAAUUUGCUGCACGCGCCGUGC-UGAAAAGUUAAGACGGCGGAGGGGA ..-------...(((((.....(((.((((((((....((((.((((.(((((.....)))))...((((((.........)))))).)))).)-))).....)))))))).)))))))) ( -47.50) >DroSec_CAF1 31624 119 - 1 AGCACCCAACUUUCCCCACACUUCCACCGUUUUGUUCCUCGGAAUGGGGACUCUUUAUGGGGCAGGGCGUGUUAAUUUGCUGCACGCGCCGUGC-UGAAAAGUUAAGAUGGCGGAGGGGA ....(((..(((.(((((....(((.((((((((.....))))))))))).......))))).)))((((((.........))))))(((((((-(....)))....)))))...))).. ( -44.00) >DroSim_CAF1 33467 119 - 1 AGCACCCAACUUUCCCCACAUUUCCAACGUUUUGUUCCUCGGAAUGGGGACUCUUUAUGGGGCAGGGCGUGUUAAUUUGCUGCACGCGCCGUGC-UGAAAAGUUAAGACGGCGGAGGGGA ....(((..(((.(((((....(((..(((((((.....))))))).))).......))))).)))((((((.........))))))(((((((-(....)))....)))))...))).. ( -41.30) >DroEre_CAF1 31108 119 - 1 AGCCCCCAGUUUUCCCCACAUCU-CCCCGUUUUGUUCUUCGGGAUGGGGACUCUUUAUGAGGCAGGGCGUGUUAAUUUGUUGCACGCGCCGGGCAUGAAAAGUUAAGACGGCGGCGGGGA ............(((((.....(-((((((((((.....)))))))))))(((.....)))((...((((((.........))))))((((..(.(((....))).).)))).))))))) ( -44.00) >DroYak_CAF1 68167 120 - 1 AGCCCCCAACUUUCCCCACAUUUUCCCAGUUUUGUUCUAAGGAAUGGGGACUCUUUAUGAGGCAGGGUGUGUUAAUUUGGUACACGCGCUGGGCAUGAAAAGUUAAGAUGGGGGAGGGGA ...((((.....(((((.((((((((((.((((......)))).))))((((.((((((...(((.((((((.........)))))).)))..)))))).))))))))))))))))))). ( -47.00) >consensus AGCACCCAACUUUCCCCACAUUUCCACCGUUUUGUUCCUCGGAAUGGGGACUCUUUAUGAGGCAGGGCGUGUUAAUUUGCUGCACGCGCCGUGC_UGAAAAGUUAAGACGGCGGAGGGGA ............(((((.....(((.(((((((((((....))))...((((.((((....(((.(((((((((......)).))))))).))).)))).))))))))))).)))))))) (-33.94 = -34.26 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:29 2006