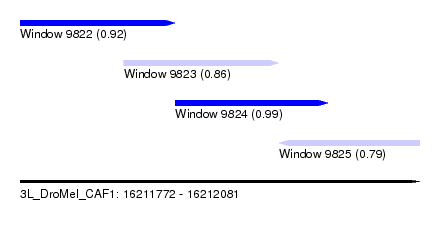
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,211,772 – 16,212,081 |
| Length | 309 |
| Max. P | 0.992798 |

| Location | 16,211,772 – 16,211,892 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -31.59 |
| Consensus MFE | -26.40 |
| Energy contribution | -27.80 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16211772 120 + 23771897 CCGACACACAGGCUGCAUAUCUAAUGAGCAUCGGAUACAGAUUCAUCUCGAGUGGCACUUGGCCCAAUAUUGCACAGCGAUUGCCAUUCUCCCACUUGAUUGUUCUCACGUAGACCGAUA ...........(((.(((.....))))))(((((.((((((..((..((((((((....((((.....(((((...))))).)))).....)))))))).)).)))...)))..))))). ( -31.20) >DroSec_CAF1 30475 120 + 1 CCGACACACAGGCUGCAUUUCUAAUGAGCAUCGGAUACACAUCCAUCUCGAGUGGCACUUGGCCCAAUAUUGCACAGCGAUUGCCAUUCUGCCACUUGAUUGUUCUCACGUAGACCGAUA ..........((((((.........(((((..((((....))))...((((((((((..((((.....(((((...))))).))))...)))))))))).)))))....)))).)).... ( -36.42) >DroSim_CAF1 32358 120 + 1 CCGACACACAGGCUGCAUAUCUAAUGAGCAUCGGAUACAGAUCCAUCUCGAGUGGCACUUGGCCCAAUAUUGCACAGCGAUUGCCAUUCUGCCACUUGAUUGUUCUCACGUAGACCGAUA ..........((((((.........(((((..((((....))))...((((((((((..((((.....(((((...))))).))))...)))))))))).)))))....)))).)).... ( -36.72) >DroEre_CAF1 30040 120 + 1 CCAACACACAGGCUGCAUAUCUAAUGAGAAUCGGAUACAGAUCCAUCUCGAGUGGCACUUCACCCAAUACUGCACAGCAAUUGCCAAUCUGCCUCUUGAUUGUUCCCACGUACACCGAUA ..........((..(((.(((....((((...((((....)))).))))(((.((((.......((((..(((...)))))))......)))).)))))))))..))............. ( -22.12) >DroYak_CAF1 67119 120 + 1 CCGACACACAGGCUGCAUAGCUAAUGAGAAUCGGAUACAGAUCCAUCUCGAGUGGCACUUCACCCAAUAUUGCACAGCAAUUGCCUAUCUGCCACUUGAUUGUUCUCACGUAGACCGAUA ..........((((((........(((((((.((((....))))...((((((((((...........(((((...)))))........))))))))))..))))))).)))).)).... ( -31.51) >consensus CCGACACACAGGCUGCAUAUCUAAUGAGCAUCGGAUACAGAUCCAUCUCGAGUGGCACUUGGCCCAAUAUUGCACAGCGAUUGCCAUUCUGCCACUUGAUUGUUCUCACGUAGACCGAUA ..........((((((................((((....))))...((((((((((..((((.....(((((...))))).))))...))))))))))..........)))).)).... (-26.40 = -27.80 + 1.40)


| Location | 16,211,852 – 16,211,972 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -28.84 |
| Energy contribution | -29.08 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16211852 120 + 23771897 UUGCCAUUCUCCCACUUGAUUGUUCUCACGUAGACCGAUAUUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGUGCCUGCCGCAUAUAAAGUUGGCAGUAGCCAGCGAUUUAAUUCGAACCA .....................((((.(((...((((.(....).))))...))).(((((((.....(((((....))))).......((((((....))))))...))))))))))).. ( -32.70) >DroSec_CAF1 30555 120 + 1 UUGCCAUUCUGCCACUUGAUUGUUCUCACGUAGACCGAUAUUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGUGCCUGCCGCAUAUAAAGUUGGCAGUAGCCAACGAUUUAAUUCGAACCA ..((....((((((((((...((((.(((...((((.(....).))))...))).))))...)....(((((....))))).....))).))))))..))...(((......)))..... ( -34.30) >DroSim_CAF1 32438 120 + 1 UUGCCAUUCUGCCACUUGAUUGUUCUCACGUAGACCGAUAUUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGUGCCUGCCGCAUAUAAAGUUGGCAGUAGCCAACGAUUUAAUUCGAACCA ..((....((((((((((...((((.(((...((((.(....).))))...))).))))...)....(((((....))))).....))).))))))..))...(((......)))..... ( -34.30) >DroEre_CAF1 30120 116 + 1 UUGCCAAUCUGCCUCUUGAUUGUUCCCACGUACACCGAUAUUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGUGCCUGCCGCAUAU----UUGGCAGUAGCCAAAGAUUUGAUUCGGGCCA ....(((((........)))))..(((..(((((((.(....).)))....))))(((((((.((...((((..((((((.....----.))))))..))))...))))))))))))... ( -34.70) >DroYak_CAF1 67199 116 + 1 UUGCCUAUCUGCCACUUGAUUGUUCUCACGUAGACCGAUAUUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGCGCCUGCCGCAUAU----UUGGCAGUAGCGAAAGAUUUAAUGCCAACCA ..((..((((((((((((...((((.(((...((((.(....).))))...))).)))).....))))))))((((((((.....----.))))))..))...)))).....))...... ( -36.90) >consensus UUGCCAUUCUGCCACUUGAUUGUUCUCACGUAGACCGAUAUUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGUGCCUGCCGCAUAUAAAGUUGGCAGUAGCCAACGAUUUAAUUCGAACCA ..((....(((((...((...((((.(((...((((.(....).))))...))).))))...))...(((((....)))))..........)))))..)).................... (-28.84 = -29.08 + 0.24)



| Location | 16,211,892 – 16,212,010 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.23 |
| Mean single sequence MFE | -37.36 |
| Consensus MFE | -31.44 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16211892 118 + 23771897 UUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGUGCCUGCCGCAUAUAAAGUUGGCAGUAGCCAGCGAUUUAAUUCGAACCAAAAAUAGCAGCUCGUGUUGCUCAAGGACCGA--AGGACUC (((.((((...((.((((((((.....(((((....))))).......((((((....))))))...)))))))).)).......((((((....))))))....)))).)--))..... ( -38.40) >DroSec_CAF1 30595 118 + 1 UUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGUGCCUGCCGCAUAUAAAGUUGGCAGUAGCCAACGAUUUAAUUCGAACCAAAAAUAGCAGCUCGUGUUGCCCAAGGACCGA--AGGACUC (((.((((...((.((((((((.....(((((....))))).......((((((....))))))...)))))))).))........(((((....))))).....)))).)--))..... ( -37.50) >DroSim_CAF1 32478 118 + 1 UUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGUGCCUGCCGCAUAUAAAGUUGGCAGUAGCCAACGAUUUAAUUCGAACCAAAAAUAGCAGCUCGUGUUGCCCAAGGAGCGA--AGGACUC (((((......((.((((((((.....(((((....))))).......((((((....))))))...)))))))).))........(((((....))))))))))((((..--..).))) ( -36.50) >DroEre_CAF1 30160 114 + 1 UUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGUGCCUGCCGCAUAU----UUGGCAGUAGCCAAAGAUUUGAUUCGGGCCAAAAAUGGCAGCUCGUGUUGCCCAAGGACCAA--AGGACUC ....((((...((.((((((((.((...((((..((((((.....----.))))))..))))...)))))))))).)).......((((((....))))))....))))..--....... ( -41.50) >DroYak_CAF1 67239 116 + 1 UUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGCGCCUGCCGCAUAU----UUGGCAGUAGCGAAAGAUUUAAUGCCAACCAAAAAUAGCAGCUCGUGUUGUCCAAGGACCAAAAAGGACUC ...(((((..((..(((.((.((....(((((....)))))....----((((((....(....)......)))))).........)))).)))..))(((....))).......))))) ( -32.90) >consensus UUUGGGUCAAAGUGCGAAUUAGCAUAAGUGGUGCCUGCCGCAUAUAAAGUUGGCAGUAGCCAACGAUUUAAUUCGAACCAAAAAUAGCAGCUCGUGUUGCCCAAGGACCGA__AGGACUC ....((((...((.((((((((.((...((((..((((((..........))))))..))))...)))))))))).))........(((((....))))).....))))........... (-31.44 = -31.60 + 0.16)


| Location | 16,211,972 – 16,212,081 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.82 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -21.91 |
| Energy contribution | -22.27 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16211972 109 - 23771897 UAAUGGCACUCGUCCUGUAAUUUCAUUUUUCAUGUAAGAC-------CAGUGGGG--AGAAAAUUUAAUGAAAUUUUCAAGAGUCCU--UCGGUCCUUGAGCAACACGAGCUGCUAUUUU .((((((((((((..(((..(..((((..((......)).-------.))))..)--.((((((((....))))))))..(((..(.--...)..)))..)))..))))).))))))).. ( -28.60) >DroSec_CAF1 30675 110 - 1 UAAUGGUACUCGUCCUGUAAUUUCAUUUUUCAUGUAAGAC-------CAGUGGGG-GGGUAAAUUUAAUGAAAUUUUCAAGAGUCCU--UCGGUCCUUGGGCAACACGAGCUGCUAUUUU .((((((((((((..((((((((((((.......((...(-------(.....))-...)).....))))))))).(((((.(.((.--..)).)))))))))..))))).))))))).. ( -25.50) >DroSim_CAF1 32558 111 - 1 UAAUGGCACUCGUCCUGUAAUUUCAUUUUUCAUGUAAGAC-------CAGUGGGGCGGGGAAAUUUAAUGAAAUUUUCAAGAGUCCU--UCGCUCCUUGGGCAACACGAGCUGCUAUUUU .((((((((((((..(((...........((......))(-------(((.((((((((((.......(((.....)))....))).--))))))))))))))..))))).))))))).. ( -33.70) >DroEre_CAF1 30236 108 - 1 UAAUGGCACUCGUUCUGUUAUUUCAUUUUUCAUAUAACAC-------CAGUAGG---GGAAAAUUUAAUGAAAUUUUCAAGAGUCCU--UUGGUCCUUGGGCAACACGAGCUGCCAUUUU .((((((((((((...(((((............)))))((-------(((.(((---(((((((((....)))))))).....))))--))))).....(....)))))).))))))).. ( -28.40) >DroYak_CAF1 67315 117 - 1 UAAUGGCACUCACUCUGUAAUUUGAUUUUUCAUGUAACACCAGUCACCAGUAGG---GGAAAAUUUAAUGAAAUUUUCAAGAGUCCUUUUUGGUCCUUGGACAACACGAGCUGCUAUUUU .((((((((((....(((.((.(((....))).)).)))((((..(((((.(((---(((((((((....))))))))......)))).)))))..)))).......))).))))))).. ( -29.30) >consensus UAAUGGCACUCGUCCUGUAAUUUCAUUUUUCAUGUAAGAC_______CAGUGGGG__GGAAAAUUUAAUGAAAUUUUCAAGAGUCCU__UCGGUCCUUGGGCAACACGAGCUGCUAUUUU .((((((((((((..((((((((((((.......................................))))))))).(((((.(.((.....)).)))))))))..))))).))))))).. (-21.91 = -22.27 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:27 2006