
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,210,287 – 16,210,519 |
| Length | 232 |
| Max. P | 0.990583 |

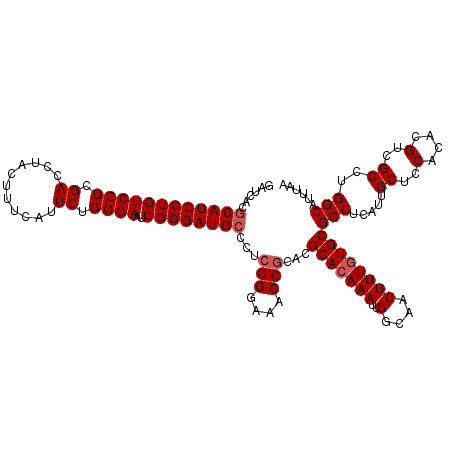
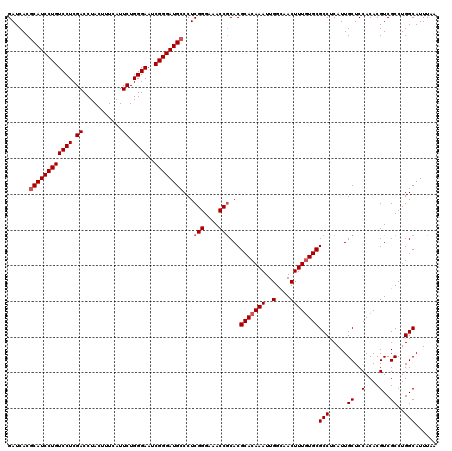
| Location | 16,210,287 – 16,210,407 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.58 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -32.78 |
| Energy contribution | -33.38 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16210287 120 - 23771897 GAUCACGCAUCCUGUCCUCGACCUACUUUUAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCGCACGCACAAAUUGGUAACUUUGUGCGCCUCAUUGCUCCACACGUCGCCUGGCAUUUAA ......((((((((((((.((...........)).))))..))))))))...(((....)))...(((((((........)))))))(((.....((..(....)..))..)))...... ( -35.40) >DroSec_CAF1 29109 120 - 1 GAUCACGCAUCCUGUCCUCGACCUACUUUCAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCACACGCACAAAUUGGCAACUUUUUGCGCCCCAUUGCUCCACACGUCGCCUGGCAUUUAA ......((((((((((((.((...........)).))))..))))))))....((....))....((.((((..(....)..)))).)).(((..((..(....)..)).)))....... ( -31.60) >DroSim_CAF1 30980 120 - 1 GAUCACGCAUCCUGUCCUCGACCUACUUUCAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCGCACGCACAAAUUGGCAACUUUGUGCGCCCCAUUGCUCCACACGUCGCCUGGCAUUUAA ......((((((((((((.((...........)).))))..))))))))...(((....)))...(((((((..(....))))))))(((.....((..(....)..))..)))...... ( -36.00) >DroEre_CAF1 28615 120 - 1 GAUCACGCAUCCUGUCCUCGACCUACUUUCAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCGCACGCACAAAUUGGCAACUUUGUGCGCCUCAUUGCUCCACACGUCGCCUGGCAUUUAA ......((((((((((((.((...........)).))))..))))))))...(((....)))...(((((((..(....))))))))(((.....((..(....)..))..)))...... ( -36.70) >DroYak_CAF1 65694 120 - 1 GAUCACACAUCCUGUCCUCGACCUACUUUAAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCGCACGCACAAAUUGGCAACUUUGUGCGCCUCAUUGCUCCACACGUCGCCUGGCAUUUAA .......(((((((((((.((...........)).))))..)))))))....(((....)))...(((((((..(....))))))))(((.....((..(....)..))..)))...... ( -33.10) >consensus GAUCACGCAUCCUGUCCUCGACCUACUUUCAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCGCACGCACAAAUUGGCAACUUUGUGCGCCUCAUUGCUCCACACGUCGCCUGGCAUUUAA ......((((((((((((.((...........)).))))..))))))))...(((....)))...(((((((..(....))))))))(((.....((..(....)..))..)))...... (-32.78 = -33.38 + 0.60)

| Location | 16,210,327 – 16,210,447 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.25 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -34.06 |
| Energy contribution | -34.74 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16210327 120 - 23771897 AUAAUUGCCAGAAGAAGCCGUCCAAGUGCCGCACCCACUGGAUCACGCAUCCUGUCCUCGACCUACUUUUAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCGCACGCACAAAUUGGUAAC ....(((((((.....((.(((((.(((.......))))))))...((((((((((((.((...........)).))))..))))))))...(((....)))...)).....))))))). ( -35.80) >DroSec_CAF1 29149 120 - 1 AUAAUUGCCAGAAGAAGCCGUCCAAGUGCCGCACCCACUGGAUCACGCAUCCUGUCCUCGACCUACUUUCAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCACACGCACAAAUUGGCAAC ....(((((((.....((.(((((.(((.......))))))))...((((((((((((.((...........)).))))..))))))))....((....))....)).....))))))). ( -36.90) >DroSim_CAF1 31020 120 - 1 AUAAUUGCCAGAAGAAGCCGUCCAAGUGCCGCACCCACUGGAUCACGCAUCCUGUCCUCGACCUACUUUCAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCGCACGCACAAAUUGGCAAC ....(((((((.....((.(((((.(((.......))))))))...((((((((((((.((...........)).))))..))))))))...(((....)))...)).....))))))). ( -37.80) >DroEre_CAF1 28655 120 - 1 AUAAUUGCCACAAGAAGCCGUCCAAGUGCCGCACCCACUGGAUCACGCAUCCUGUCCUCGACCUACUUUCAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCGCACGCACAAAUUGGCAAC ....((((((......((.(((((.(((.......))))))))...((((((((((((.((...........)).))))..))))))))...(((....)))...))......)))))). ( -37.00) >DroYak_CAF1 65734 120 - 1 AUAAUUGCUCCAAGAAGCCGUCCAAGUGCCGCACCCACUGGAUCACACAUCCUGUCCUCGACCUACUUUAAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCGCACGCACAAAUUGGCAAC ................((((.....((((.((((((...((((.....))))..((((.((...........)).))))...))).)))...(((....)))...))))....))))... ( -32.00) >consensus AUAAUUGCCAGAAGAAGCCGUCCAAGUGCCGCACCCACUGGAUCACGCAUCCUGUCCUCGACCUACUUUCAUUCUGGGAAUCGGGAUGCCCUCGGGAAACCGCACGCACAAAUUGGCAAC ....(((((((.....((.(((((.(((.......))))))))...((((((((((((.((...........)).))))..))))))))...(((....)))...)).....))))))). (-34.06 = -34.74 + 0.68)

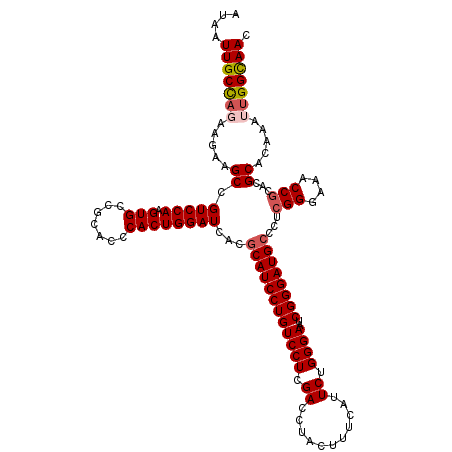

| Location | 16,210,407 – 16,210,519 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -34.68 |
| Energy contribution | -35.16 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16210407 112 + 23771897 CAGUGGGUGCGGCACUUGGACGGCUUCUUCUGGCAAUUAUGAAUGAAAUCCUUUCAGGAGCGAAAUCCGUUCCACCGGCAGUCGCCCAAGGUCGCUCCAAAGCUUGGU--------CUCU .(((.((.(((((.(((((.(((((....((((......((((.(.....).))))((((((.....)))))).)))).))))).)))))))))).))...)))....--------.... ( -41.60) >DroSec_CAF1 29229 112 + 1 CAGUGGGUGCGGCACUUGGACGGCUUCUUCUGGCAAUUAUGAAUGAAAUCCUUUCAGGAGCGAAAUCCGUUCCACCGGCAGUCACCCAAGGUCGCUCCAAAGCUUGGU--------CUCU .(((.((.(((((.(((((..((((....((((......((((.(.....).))))((((((.....)))))).)))).))))..)))))))))).))...)))....--------.... ( -38.50) >DroSim_CAF1 31100 112 + 1 CAGUGGGUGCGGCACUUGGACGGCUUCUUCUGGCAAUUAUGAAUGAAAUCCUUUCAGGAGCGAAAUCCGUUCCACCGGCAGUCGCCCAAGGUCGCUCCAAAGCUUGGU--------CUCU .(((.((.(((((.(((((.(((((....((((......((((.(.....).))))((((((.....)))))).)))).))))).)))))))))).))...)))....--------.... ( -41.60) >DroEre_CAF1 28735 120 + 1 CAGUGGGUGCGGCACUUGGACGGCUUCUUGUGGCAAUUAUGAAUGAAAUCCUUUCAGGAGCGAAAUCCGCUCCACCGGCAGUCGCCCAAGGUCGCUCGCUCGCUCGGUCGGUCCGUCGCU .((((((((((((.(((((.(((((.((.((........((((.(.....).))))((((((.....)))))))).)).))))).))))))))))..)))))))(((.....)))..... ( -47.00) >DroYak_CAF1 65814 112 + 1 CAGUGGGUGCGGCACUUGGACGGCUUCUUGGAGCAAUUAUGAAUGAAAUCCUUUCAGGAGCGAAAUCCGUUCCACCGGCAGUUGCCCAAGGUCGCUCGGUCUCUCGGU--------CUCU ......(.(((((.(((((.(((((.((((((((.....((((.(.....).))))(((......)))))))))..)).))))).)))))))))).)(..(....)..--------)... ( -36.80) >consensus CAGUGGGUGCGGCACUUGGACGGCUUCUUCUGGCAAUUAUGAAUGAAAUCCUUUCAGGAGCGAAAUCCGUUCCACCGGCAGUCGCCCAAGGUCGCUCCAAAGCUUGGU________CUCU .(((.((.(((((.(((((.(((((.((...........((((.(.....).))))((((((.....))))))...)).))))).)))))))))).))...)))................ (-34.68 = -35.16 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:20 2006