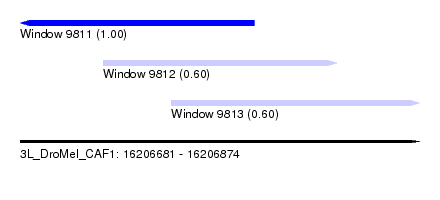
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,206,681 – 16,206,874 |
| Length | 193 |
| Max. P | 0.995081 |

| Location | 16,206,681 – 16,206,794 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.78 |
| Mean single sequence MFE | -38.49 |
| Consensus MFE | -33.44 |
| Energy contribution | -33.80 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16206681 113 - 23771897 GGCCCAAAACGCCGAGCUGCAGCCUGCAGUUGCGCUCUCUGCGGAGAGAAAGUAGCUAGUAAGAGAGA-------AGGUUGUCGGUGUGCGUGAAUUUCGGAAGGAUGUGGUAGAAAGUA .(((((..((((...((((((((((((((((((.(((((....)))))...)))))).))........-------)))))).))))..))))....(((....))))).)))........ ( -38.00) >DroSec_CAF1 25565 113 - 1 GGCCCAAAACGCCGAGCUGCAGCCUGCAGUUGCGCUCUCUGCGGAGAGAAAGUAGCUAGUAAGAGAGA-------AGGUUGUCGGUGUGCGUGAAUUUCGGAAGGAUGUGGUAGAAAGUA .(((((..((((...((((((((((((((((((.(((((....)))))...)))))).))........-------)))))).))))..))))....(((....))))).)))........ ( -38.00) >DroSim_CAF1 27442 113 - 1 GGCCCAAAACGCCGAGCUGCAGCCUGCAGUUGCGCUCUCUGCGGAGAGAAAGUAGCUAGUAAGAGAGA-------AGGUUGUCGGUGUGCGUGAAUUUCGGAAGGAUGUGGUAUAAAGUA .(((((..((((...((((((((((((((((((.(((((....)))))...)))))).))........-------)))))).))))..))))....(((....))))).)))........ ( -38.00) >DroEre_CAF1 25070 119 - 1 GGCCCAAAACGCCGAGCUGCAGCCUGCAGUUGCGCUCUCCGCG-AGAGAAAGUAGCUAGUAAGAGAGAGAGAGAGAGGCUGUCGGUGUGCGUGAAUUUCGGAAGGAUGUGGCAGAAAGUA .(((((..((((...((((((((((((((((((.(((((...)-))))...)))))).))...............)))))).))))..))))....(((....))))).)))........ ( -39.86) >DroYak_CAF1 61967 112 - 1 GGCCCACAACGGCGAGCUGCAGUCUGCAGUUGCGCUCUCAGCG-AGAGAAAGUAGCUAGUAAGAGAGA-------AGGUUGUCGGUGUGCGUAAAUUUCGGAAGGAUGUGGGAGAAAGUA ..((((((.((((((.((.(..(((((((((((.(((((...)-))))...)))))).)).)))..).-------)).))))))............(((....)))))))))........ ( -38.60) >consensus GGCCCAAAACGCCGAGCUGCAGCCUGCAGUUGCGCUCUCUGCGGAGAGAAAGUAGCUAGUAAGAGAGA_______AGGUUGUCGGUGUGCGUGAAUUUCGGAAGGAUGUGGUAGAAAGUA .(((((..((((...((((((((((((((((((.(((((....)))))...)))))).))...............)))))).))))..))))....(((....))))).)))........ (-33.44 = -33.80 + 0.36)

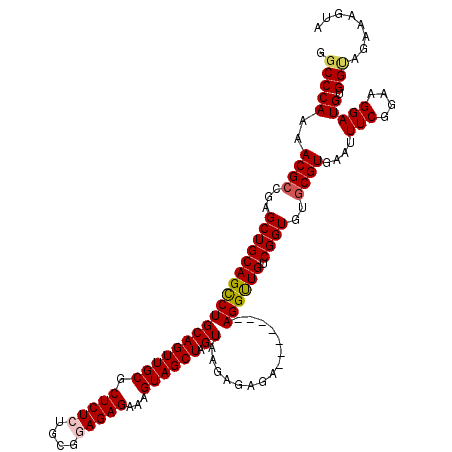
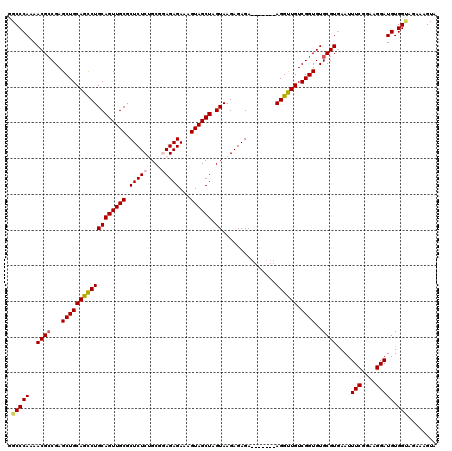
| Location | 16,206,721 – 16,206,834 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.78 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -36.10 |
| Energy contribution | -36.86 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16206721 113 + 23771897 AACCU-------UCUCUCUUACUAGCUACUUUCUCUCCGCAGAGAGCGCAACUGCAGGCUGCAGCUCGGCGUUUUGGGCCAAUGAACUCGGGCUGCAAGGACAUUGCAGUUGCCGUCGAG .....-------...(((..............(((((....))))).((((((((((.(((((((((((.((((.........)))))))))))))..))...))))))))))....))) ( -39.60) >DroSec_CAF1 25605 113 + 1 AACCU-------UCUCUCUUACUAGCUACUUUCUCUCCGCAGAGAGCGCAACUGCAGGCUGCAGCUCGGCGUUUUGGGCCAAUGAACUCGGGCUGCAAGGACAUUGCAGUUGCCGUCGAG .....-------...(((..............(((((....))))).((((((((((.(((((((((((.((((.........)))))))))))))..))...))))))))))....))) ( -39.60) >DroSim_CAF1 27482 113 + 1 AACCU-------UCUCUCUUACUAGCUACUUUCUCUCCGCAGAGAGCGCAACUGCAGGCUGCAGCUCGGCGUUUUGGGCCAAUGAACUCGGGCUGCAAGGACAUUGCAGUUGCUGUCGAG .....-------...(((..............(((((....))))).((((((((((.(((((((((((.((((.........)))))))))))))..))...))))))))))....))) ( -40.00) >DroEre_CAF1 25110 119 + 1 AGCCUCUCUCUCUCUCUCUUACUAGCUACUUUCUCU-CGCGGAGAGCGCAACUGCAGGCUGCAGCUCGGCGUUUUGGGCCAAUGAACUCGUGCUGCAAGGACAUUGCAGUUGCCGUCGAG ...(((.......((((((.....((..........-.)))))))).((((((((((.(((((((.(((.((((.........))))))).)))))..))...))))))))))....))) ( -36.90) >DroYak_CAF1 62007 112 + 1 AACCU-------UCUCUCUUACUAGCUACUUUCUCU-CGCUGAGAGCGCAACUGCAGACUGCAGCUCGCCGUUGUGGGCCAAUGAACUGGAGCUGCAAGGACAUUGCAGUUGCCGUCGAG .....-------...(((.............((((.-....))))(.((((((((((.((((((((((((......)))((......))))))))).))....)))))))))))...))) ( -38.50) >consensus AACCU_______UCUCUCUUACUAGCUACUUUCUCUCCGCAGAGAGCGCAACUGCAGGCUGCAGCUCGGCGUUUUGGGCCAAUGAACUCGGGCUGCAAGGACAUUGCAGUUGCCGUCGAG ...............(((......(((.((((........)))))))((((((((((.(((((((((((.((((.........))))))))))))).))....))))))))))....))) (-36.10 = -36.86 + 0.76)

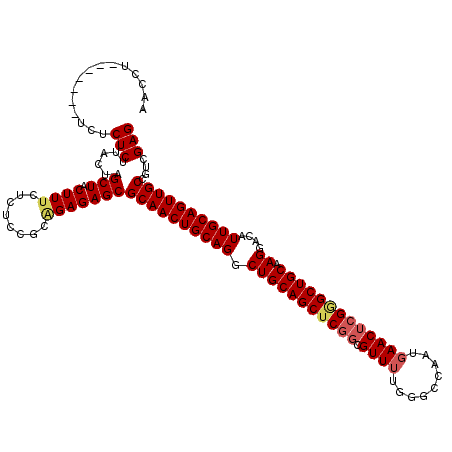
| Location | 16,206,754 – 16,206,874 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.81 |
| Mean single sequence MFE | -42.23 |
| Consensus MFE | -36.24 |
| Energy contribution | -36.12 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16206754 120 + 23771897 AGAGAGCGCAACUGCAGGCUGCAGCUCGGCGUUUUGGGCCAAUGAACUCGGGCUGCAAGGACAUUGCAGUUGCCGUCGAGUGACAUUACGAAAAGCAAGAACAUCAGCAGGCGGGCAGGA .....((((((((((((.(((((((((((.((((.........)))))))))))))..))...))))))))))((((..(((....))).....((..........)).)))).)).... ( -41.00) >DroSec_CAF1 25638 120 + 1 AGAGAGCGCAACUGCAGGCUGCAGCUCGGCGUUUUGGGCCAAUGAACUCGGGCUGCAAGGACAUUGCAGUUGCCGUCGAGUGACAUUACGAAAGGCAAGAACAGCAGCAGGCAGGCAGGA .......((..((((..(((((.((((((((...((((........))))((((((((.....))))))))..)))))))).......(....).........)))))..)))))).... ( -43.90) >DroSim_CAF1 27515 120 + 1 AGAGAGCGCAACUGCAGGCUGCAGCUCGGCGUUUUGGGCCAAUGAACUCGGGCUGCAAGGACAUUGCAGUUGCUGUCGAGUGACAUUACGAAAGGCAAGAACAGCAGCAGGCAGGCAGGA .......((..((((..(((((.(.((.((.(((((....((((.(((((((((((((.....))))))).....))))))..)))).))))).))..)).).)))))..)))))).... ( -42.90) >DroEre_CAF1 25149 116 + 1 GGAGAGCGCAACUGCAGGCUGCAGCUCGGCGUUUUGGGCCAAUGAACUCGUGCUGCAAGGACAUUGCAGUUGCCGUCGAGUGACAUUACGAAAGGCAGGAACAGCGGCAGGCGGG----A ...........((((..(((((.(.((.((.(((((....((((.(((((.(((((((.....)))))))......)))))..)))).))))).))..)).).)))))..)))).----. ( -41.10) >DroYak_CAF1 62039 116 + 1 UGAGAGCGCAACUGCAGACUGCAGCUCGCCGUUGUGGGCCAAUGAACUGGAGCUGCAAGGACAUUGCAGUUGCCGUCGAGUGACAUUACGAAAGGCAAGACCAGCGGCAGGCAGG----A .....((((((((((((.((((((((((((......)))((......))))))))).))....)))))))))).((((..........(....)..........))))..))...----. ( -42.25) >consensus AGAGAGCGCAACUGCAGGCUGCAGCUCGGCGUUUUGGGCCAAUGAACUCGGGCUGCAAGGACAUUGCAGUUGCCGUCGAGUGACAUUACGAAAGGCAAGAACAGCAGCAGGCAGGCAGGA ...........((((..(((((.((((((....)))))).((((.(((((((((((((.....))))))).....))))))..))))................)))))..))))...... (-36.24 = -36.12 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:15 2006