
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,202,350 – 16,202,469 |
| Length | 119 |
| Max. P | 0.995721 |

| Location | 16,202,350 – 16,202,469 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
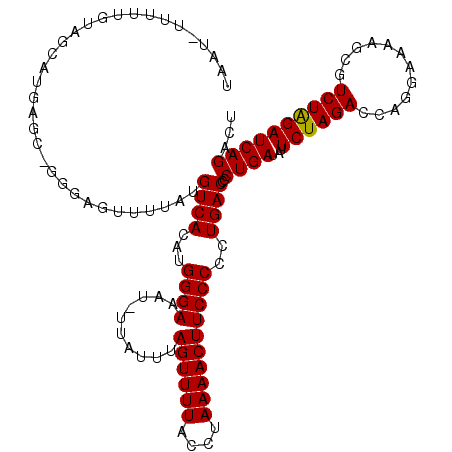
>3L_DroMel_CAF1 16202350 119 + 23771897 UAAUCUUUUUGUAGCAUGAGCUGGGAGUUUUAUGUCACAAGGGAAUU-UUUUUUAGUUUUACCUAAAACUUCCCACUGACCCCUGAUAUUCUAGACCAGGAAAAGCAUCUGGAUCAGCAU .............((.(((.(..(..(((((..((((...(((((..-..((((((......)))))).)))))..)))).((((...........)))).)))))..)..).))))).. ( -27.70) >DroSec_CAF1 21314 118 + 1 UAAUGUUUUUGUAGCAUGAGCAGGGAGUUUUAUGUCAAAUGGGAAAC--UAUUUAGUUUUACCUAAAACUUCCCCCUGACCCCUGAUAUUCUAGAACAGGAAAAGCGUCUAGAUCAGACU ..(((((.....)))))...((((((((((((.(((((((((....)--))))).)....)).)))))))...)))))....((((...((((((...(......).))))))))))... ( -28.70) >DroSim_CAF1 23234 98 + 1 ----------------------GGGAGUUUUAUGUCACAUGGGAAAUUUUAUUUAGUUUUACCAAAAACUUCCCCCUGACCUCUGAUAUUCUAGACCAGGAAAAGCGUCUAGAUCAGACU ----------------------(((((((((........(((((((((......)))))).))))))))))))........(((((...((((((((.......).)))))))))))).. ( -24.30) >consensus UAAU_UUUUUGUAGCAUGAGC_GGGAGUUUUAUGUCACAUGGGAAAU_UUAUUUAGUUUUACCUAAAACUUCCCCCUGACCCCUGAUAUUCUAGACCAGGAAAAGCGUCUAGAUCAGACU .................................((((...((((..........((((((....))))))))))..))))..((((...((((((............))))))))))... (-17.35 = -17.13 + -0.22)

| Location | 16,202,350 – 16,202,469 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -20.43 |
| Energy contribution | -21.43 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16202350 119 - 23771897 AUGCUGAUCCAGAUGCUUUUCCUGGUCUAGAAUAUCAGGGGUCAGUGGGAAGUUUUAGGUAAAACUAAAAAA-AAUUCCCUUGUGACAUAAAACUCCCAGCUCAUGCUACAAAAAGAUUA ..((((((((.((......))(((((.......)))))))))))))(((((.((((((......))))))..-..)))))..................(((....)))............ ( -28.80) >DroSec_CAF1 21314 118 - 1 AGUCUGAUCUAGACGCUUUUCCUGUUCUAGAAUAUCAGGGGUCAGGGGGAAGUUUUAGGUAAAACUAAAUA--GUUUCCCAUUUGACAUAAAACUCCCUGCUCAUGCUACAAAAACAUUA (((.(((((((((.((.......))))))))....(((((((((((((((((.(((((......)))))..--.)))))).))))))........))))).))).)))............ ( -31.50) >DroSim_CAF1 23234 98 - 1 AGUCUGAUCUAGACGCUUUUCCUGGUCUAGAAUAUCAGAGGUCAGGGGGAAGUUUUUGGUAAAACUAAAUAAAAUUUCCCAUGUGACAUAAAACUCCC---------------------- ..((((((((((((.(.......))))))))...))))).((((.((((((((((((((.....)))...)))))))))).).))))...........---------------------- ( -30.30) >consensus AGUCUGAUCUAGACGCUUUUCCUGGUCUAGAAUAUCAGGGGUCAGGGGGAAGUUUUAGGUAAAACUAAAUAA_AUUUCCCAUGUGACAUAAAACUCCC_GCUCAUGCUACAAAAA_AUUA ...(((((((((((..........)))))))...))))..((((..((((((((((((......)))))....)))))))...))))................................. (-20.43 = -21.43 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:09 2006