
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,196,595 – 16,196,703 |
| Length | 108 |
| Max. P | 0.687040 |

| Location | 16,196,595 – 16,196,703 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -17.51 |
| Energy contribution | -18.73 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
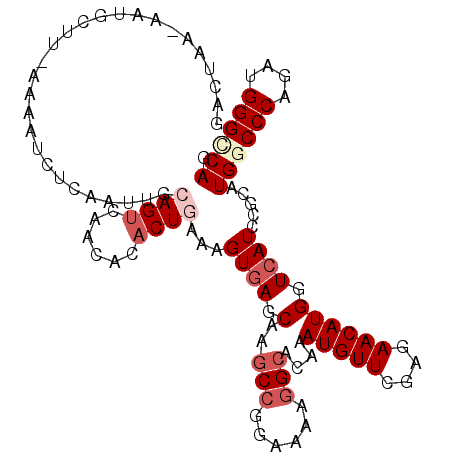
>3L_DroMel_CAF1 16196595 108 - 23771897 AGCUAAUAACUUAU-AAAAUAUCAUU--UAGUUAAUACCCUCAAAGUCAGCAAGCCGGAGAAGGCGCAAAUGUUCGAGAACAUGGUCAUCCGCAUGGCCCAGAUGGGUCAG ((((((........-..........)--)))))....................(((......)))((..(((((....)))))((....)))).((((((....)))))). ( -22.57) >DroVir_CAF1 21517 104 - 1 CGCUA---AUGU----CAAUCUCCAUUGCAGUCAACACACUGAAAGUGAGCAAGCCGGAAAAGGCGCAAAUGUUCGAGAACAUGGUCAUACGCAUGGCCCAAAUGGGCCAG .((..---((((----((.((((.....((((......)))).....(((((.(((......))).....)))))))))...))).)))..)).((((((....)))))). ( -31.30) >DroPse_CAF1 51683 111 - 1 GCUUCAGAAUGCUUAAAAAUCUCGAUUGCAGUUAACACACUGAAAGUGAGCAAGCCAGAAAAGGCCCAGAUGUUCGAGAACAUGGUCAUCCGCAUGGCCCAGAUGGGGCAG ((((((...((........((((((((((.(....).(((.....))).))))(((......)))........))))))....((((((....))))))))..)))))).. ( -30.10) >DroMoj_CAF1 18388 107 - 1 GACUA---ACACAA-ACGAUCUCAAUUGCAGUGAACACACUGAAAGUGAGCAAACCGGACAAAGCACAAAUGUUCGAGAACAUGGUCAUACGUAUGGCCCAAAUGGGCCAA (((((---......-.....((((....(((((....)))))....)))).....((((((.........))))))......))))).......((((((....)))))). ( -32.70) >DroAna_CAF1 31774 108 - 1 GAUUAAUAAAGGUU-UGUUUUCUUUU--CAGUGAACACACUCAAGGUGAGCAAGCCCGAAAAGGCACAAAUGUUCGAGAACAUGGUCAUUCGGAUGGCCCAGAUGGGUCAG ..........((((-(((((.((((.--.((((....)))).)))).)))))))))((((..(((....(((((....))))).))).))))..((((((....)))))). ( -33.50) >DroPer_CAF1 51642 111 - 1 GCUUCAGAAUGCUUCAAAAUCUCGAUUGCAGUCAACACACUGAAAGUGAGCAAGCCAGAAAAGGCCCAGAUGUUCGAGAACAUGGUCAUCCGCAUGGCCCAGAUGGGGCAG .........(((((((...((((((((((.(....).(((.....))).))))(((......)))........))))))....((((((....))))))....))))))). ( -30.40) >consensus GACUAA_AAUGCUU_AAAAUCUCAAUUGCAGUCAACACACUGAAAGUGAGCAAGCCGGAAAAGGCACAAAUGUUCGAGAACAUGGUCAUCCGCAUGGCCCAGAUGGGCCAG ............................((((......))))...((((.(..(((......)))....(((((....)))))).)))).....((((((....)))))). (-17.51 = -18.73 + 1.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:02 2006