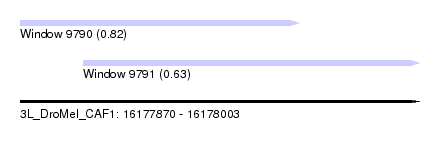
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,177,870 – 16,178,003 |
| Length | 133 |
| Max. P | 0.815722 |

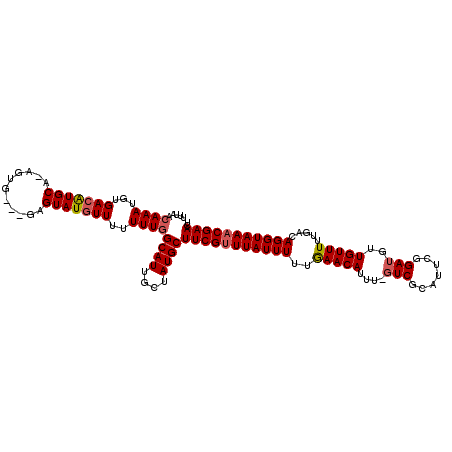
| Location | 16,177,870 – 16,177,963 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 84.89 |
| Mean single sequence MFE | -16.67 |
| Consensus MFE | -11.20 |
| Energy contribution | -11.57 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16177870 93 + 23771897 UUUCACCACUUUUUUUUUUGUUGUAACAAAUGUGACAUGCAGAGUG---GAGUAUGUUUUUUUGGCAUUGCUAUGCUUCGUUUAUUUUUGAACAUU .....(((((((......(((..((.....))..)))...))))))---)((((((((.....)))).)))).......(((((....)))))... ( -17.80) >DroSec_CAF1 35533 94 + 1 UUUCCCCACUUUUUU-UUUGUUUUAACAAAUGUGACAUGCA-AGUUUUUUUGUAUGUUUUUUUGGCAUUGCUAUGCUUCGUUUAUUUUUGAACAUU ...............-..........((((...((((((((-((....))))))))))..))))((((....))))...(((((....)))))... ( -18.60) >DroSim_CAF1 38153 86 + 1 UUUCCCCACUCUUCU------UUUAACAAAUGUGACAUGCA-AGUG---GAGUAUGUUUUUUUGGCAUUGCUAUGCUUCGUUUAUUUUUAAACAUU ...............------.....((((...(((((((.-....---..)))))))..))))((((....))))...(((((....)))))... ( -15.00) >DroEre_CAF1 35911 84 + 1 UUUCCCUCAUAUU-U------UUUAACAAAUGUGACGUGCA-AGUG---GAGUAUUUUUUUUU-GCAUUGCUAUGCUUCGCUUAUUUUUGAACAUU ......(((((((-(------......)))))))).....(-((((---((((((........-........)))))))))))............. ( -15.29) >consensus UUUCCCCACUUUUUU______UUUAACAAAUGUGACAUGCA_AGUG___GAGUAUGUUUUUUUGGCAUUGCUAUGCUUCGUUUAUUUUUGAACAUU ..........................((((...(((((((...........)))))))..))))((((....))))...(((((....)))))... (-11.20 = -11.57 + 0.38)

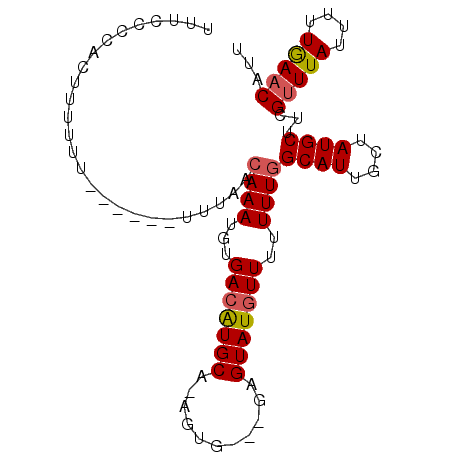
| Location | 16,177,891 – 16,178,003 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -19.75 |
| Energy contribution | -20.38 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16177891 112 + 23771897 UGUAACAAAUGUGACAUGCAGAGUG---GAGUAUGUUUUUUUGGCAUUGCUAUGCUUCGUUUAUUUUUGAACAUUUUUUCGCAUUCGGAUGUUGUUUUUGACAGGUAAACGAACU .....((((...(((((((......---..)))))))..))))((((....))))(((((((((((..(((((....((((....))))...))))).....))))))))))).. ( -25.60) >DroSec_CAF1 35553 113 + 1 UUUAACAAAUGUGACAUGCA-AGUUUUUUUGUAUGUUUUUUUGGCAUUGCUAUGCUUCGUUUAUUUUUGAACAUUU-GUCGCAUUCGGAUGUUGUUUUUGACAGGUAAACGAACU .....((((...((((((((-((....))))))))))..))))((((....))))(((((((((((...(((((((-(.......))))))))(((...)))))))))))))).. ( -29.30) >DroSim_CAF1 38168 110 + 1 UUUAACAAAUGUGACAUGCA-AGUG---GAGUAUGUUUUUUUGGCAUUGCUAUGCUUCGUUUAUUUUUAAACAUUU-GUCGCAUUCGGAUGUUGUUUUUGACAGGUAAACGAACU .....((((...(((((((.-....---..)))))))..))))((((....))))(((((((((((...(((((((-(.......))))))))(((...)))))))))))))).. ( -25.70) >DroEre_CAF1 35925 109 + 1 UUUAACAAAUGUGACGUGCA-AGUG---GAGUAUUUUUUUUU-GCAUUGCUAUGCUUCGCUUAUUUUUGAACAUUU-GUCGUGUUCAGAUGUUGUUUUUGACAGGUAAACGAACU .....((((.(..((((..(-((((---((((((........-........)))))))))))....((((((((..-...))))))))))))..).))))............... ( -24.69) >consensus UUUAACAAAUGUGACAUGCA_AGUG___GAGUAUGUUUUUUUGGCAUUGCUAUGCUUCGUUUAUUUUUGAACAUUU_GUCGCAUUCGGAUGUUGUUUUUGACAGGUAAACGAACU .....((((...(((((((...........)))))))..))))((((....))))(((((((((((..(((((....(((.......)))..))))).....))))))))))).. (-19.75 = -20.38 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:53 2006