
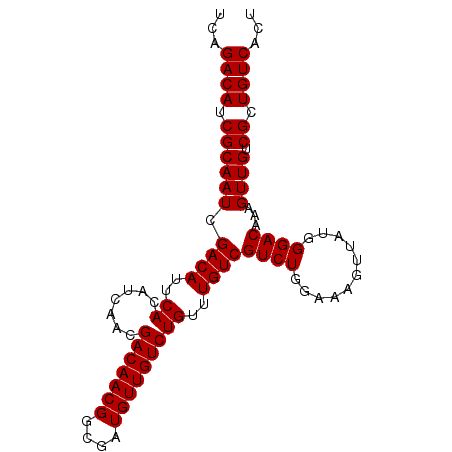
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,174,775 – 16,174,988 |
| Length | 213 |
| Max. P | 0.991892 |

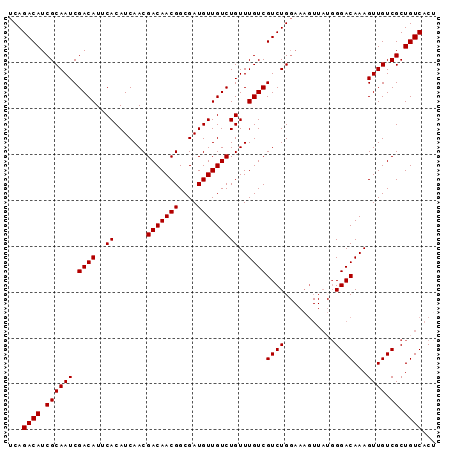
| Location | 16,174,775 – 16,174,868 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 93.73 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -23.74 |
| Energy contribution | -23.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16174775 93 - 23771897 UCAGACAUCGCAAUCGACAUUCACAUCAACGACAACGGCGAUGUUGUCUGUUUGUCGUCUGGAAAGUUAGGGGACAAAGUUGUCGCUGUCACA ...((((.((((((.((((...(((((..((....))..)))))))))..((((((.(((((....))))).)))))))))).)).))))... ( -28.60) >DroSec_CAF1 32415 83 - 1 UCAGACAUCGCAAUCGACAUUCACAUCAACGACAACGGCGAUGUUGUCUGUUUGUCGUCUG----------GGACAAAGUUGUCGCUGUCACU ...((((.((((((.((((...(((((..((....))..)))))))))..((((((.....----------.)))))))))).)).))))... ( -24.00) >DroSim_CAF1 35024 93 - 1 UCAGACAUCGCAAUCGACAUUCACAUCAACGACAACGGCGAUGUUGUCUGUUUGUCGUCUGGAAAGUUAUGGGACAAAGUUGUCGCUGUCACU ((((((..((.(((.((((...(((((..((....))..))))))))).)))))..))))))..(((.((((((((....)))).)))).))) ( -25.30) >DroEre_CAF1 32737 93 - 1 UCAGACAUCGCAAUCGACAUUCACAUCAACGACAACGGCGAUGUUGUCUGUUUGUCGUCUGGAAAGUUAUGGGACAAAGUUGUCGCUGUCACU ((((((..((.(((.((((...(((((..((....))..))))))))).)))))..))))))..(((.((((((((....)))).)))).))) ( -25.30) >consensus UCAGACAUCGCAAUCGACAUUCACAUCAACGACAACGGCGAUGUUGUCUGUUUGUCGUCUGGAAAGUUAUGGGACAAAGUUGUCGCUGUCACU ...((((.((((((.((((..((.......(((((((....)))))))))..))))((((...........))))...)))).)).))))... (-23.74 = -23.73 + -0.00)

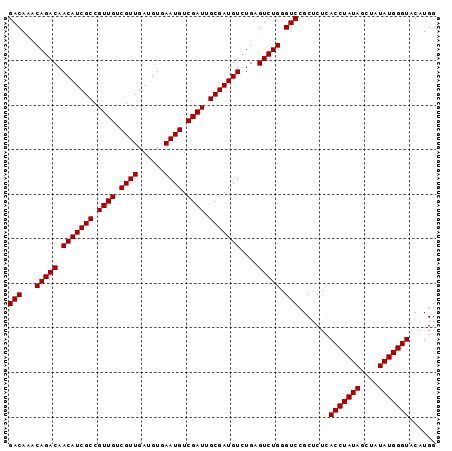
| Location | 16,174,812 – 16,174,908 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 98.44 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -33.08 |
| Energy contribution | -33.08 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16174812 96 + 23771897 GACAAACAGACAACAUCGCCGUUGUCGUUGAUGUGAAUGUCGAUUGCGAUGUCUGAGUCUGGGUCCGCUCUCACCUAUAGCUAUAUGGGUACAUGG (((...(((((.(((((((.((((.((((......)))).)))).)))))))....))))).))).......(((((((....)))))))...... ( -33.00) >DroSec_CAF1 32442 96 + 1 GACAAACAGACAACAUCGCCGUUGUCGUUGAUGUGAAUGUCGAUUGCGAUGUCUGAGUCUGGGUCCGCUCUUACCUAUAGCUAUAUGGGUACAUGG (((...(((((.(((((((.((((.((((......)))).)))).)))))))....))))).)))......((((((((....))))))))..... ( -34.00) >DroSim_CAF1 35061 96 + 1 GACAAACAGACAACAUCGCCGUUGUCGUUGAUGUGAAUGUCGAUUGCGAUGUCUGAGUCUGGGUCCGCUCUCACCUAUAGCUAUAUGGGUACAUGG (((...(((((.(((((((.((((.((((......)))).)))).)))))))....))))).))).......(((((((....)))))))...... ( -33.00) >DroEre_CAF1 32774 96 + 1 GACAAACAGACAACAUCGCCGUUGUCGUUGAUGUGAAUGUCGAUUGCGAUGUCUGAGUCUGGGUCUGCGCUCACCUAUAGCUAUAUGGGUACAUGG (((...(((((.(((((((.((((.((((......)))).)))).)))))))....))))).))).......(((((((....)))))))...... ( -33.40) >consensus GACAAACAGACAACAUCGCCGUUGUCGUUGAUGUGAAUGUCGAUUGCGAUGUCUGAGUCUGGGUCCGCUCUCACCUAUAGCUAUAUGGGUACAUGG (((...(((((.(((((((.((((.((((......)))).)))).)))))))....))))).))).......(((((((....)))))))...... (-33.08 = -33.08 + -0.00)


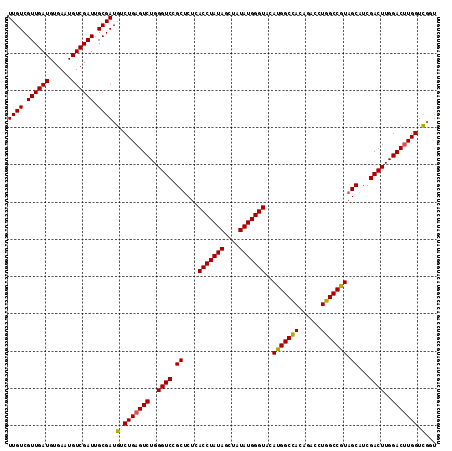
| Location | 16,174,833 – 16,174,948 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -42.11 |
| Energy contribution | -41.80 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16174833 115 + 23771897 UUGUCGUUGAUGUGAAUGUCGAUUGCGAUGUCUGAGUCUGGGUCCGCUCUCACCUAUAGCUAUAUGGGUACAUGGCCACAGACCUGGCCGUAGCAUCGACUUGGACUUGGUUGGU ((((.((((((......)))))).))))...(..((((..((((.(((...(((((((....)))))))..(((((((......))))))))))...))))..))))..)..... ( -43.10) >DroSec_CAF1 32463 115 + 1 UUGUCGUUGAUGUGAAUGUCGAUUGCGAUGUCUGAGUCUGGGUCCGCUCUUACCUAUAGCUAUAUGGGUACAUGGCCACAGACCUGGCCGUAGCAUCGACUUGGACUUGGUCGGU ((((.((((((......)))))).))))((.(..((((..((((.(((..((((((((....)))))))).(((((((......))))))))))...))))..))))..).)).. ( -46.10) >DroSim_CAF1 35082 115 + 1 UUGUCGUUGAUGUGAAUGUCGAUUGCGAUGUCUGAGUCUGGGUCCGCUCUCACCUAUAGCUAUAUGGGUACAUGGCCACAGACCUGGCCGUAGCAUCGACUUGGACUUGGUCGGU ((((.((((((......)))))).))))((.(..((((..((((.(((...(((((((....)))))))..(((((((......))))))))))...))))..))))..).)).. ( -45.10) >DroEre_CAF1 32795 115 + 1 UUGUCGUUGAUGUGAAUGUCGAUUGCGAUGUCUGAGUCUGGGUCUGCGCUCACCUAUAGCUAUAUGGGUACAUGGCUAUAGCCCUGGCCAUAGCCUCGACUUGGACCUGGUCUCC ((((.((((((......)))))).)))).....(((.(..((((((.(.(((((((((....)))))))....((((((.((....)).))))))..))).))))))..).))). ( -42.80) >consensus UUGUCGUUGAUGUGAAUGUCGAUUGCGAUGUCUGAGUCUGGGUCCGCUCUCACCUAUAGCUAUAUGGGUACAUGGCCACAGACCUGGCCGUAGCAUCGACUUGGACUUGGUCGGU ((((.((((((......)))))).)))).(.(((((((..((((.((....(((((((....)))))))..(((((((......))))))).))...))))..))))))).)... (-42.11 = -41.80 + -0.31)


| Location | 16,174,868 – 16,174,988 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -48.30 |
| Consensus MFE | -44.51 |
| Energy contribution | -44.45 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16174868 120 + 23771897 GUCUGGGUCCGCUCUCACCUAUAGCUAUAUGGGUACAUGGCCACAGACCUGGCCGUAGCAUCGACUUGGACUUGGUUGGUGGUGCCGGGUUCCUGUUCUGGUUCUGGUUGCCAAGCUCGU (((..((((.(((...(((((((....)))))))..(((((((......))))))))))...))))..)))..(.(((((...((((((..((......)).)))))).))))).).... ( -47.50) >DroSec_CAF1 32498 120 + 1 GUCUGGGUCCGCUCUUACCUAUAGCUAUAUGGGUACAUGGCCACAGACCUGGCCGUAGCAUCGACUUGGACUUGGUCGGUGGUGCCGGGUUCCUGUUCUGGUUCUGGUUGCCAAGCUCGU (((..((((.(((..((((((((....)))))))).(((((((......))))))))))...))))..)))(((((.(.(((.((((((.......)))))).))).).)))))...... ( -46.80) >DroSim_CAF1 35117 120 + 1 GUCUGGGUCCGCUCUCACCUAUAGCUAUAUGGGUACAUGGCCACAGACCUGGCCGUAGCAUCGACUUGGACUUGGUCGGUGGUGCCGGGUUCCUGUUCUGGUUCUGGUUGCCAAGCUCGU (((..((((.(((...(((((((....)))))))..(((((((......))))))))))...))))..)))(((((.(.(((.((((((.......)))))).))).).)))))...... ( -45.80) >DroEre_CAF1 32830 119 + 1 GUCUGGGUCUGCGCUCACCUAUAGCUAUAUGGGUACAUGGCUAUAGCCCUGGCCAUAGCCUCGACUUGGACCUGGUCUCCGGUGCCGGGUUCCUCUUCUGGUUCUGGUUGCCA-GCUCGU (((..((((.......(((((((....)))))))....((((((.((....)).))))))..))))..)))(((((..((((.((((((.......)))))).))))..))))-)..... ( -53.10) >consensus GUCUGGGUCCGCUCUCACCUAUAGCUAUAUGGGUACAUGGCCACAGACCUGGCCGUAGCAUCGACUUGGACUUGGUCGGUGGUGCCGGGUUCCUGUUCUGGUUCUGGUUGCCAAGCUCGU (((..((((.((....(((((((....)))))))..(((((((......))))))).))...))))..)))(((((.(.(((.((((((.......)))))).))).).)))))...... (-44.51 = -44.45 + -0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:49 2006