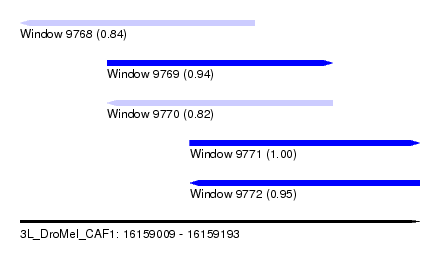
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,159,009 – 16,159,193 |
| Length | 184 |
| Max. P | 0.998659 |

| Location | 16,159,009 – 16,159,117 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -24.61 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.85 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16159009 108 - 23771897 ----------AUAUACAUAUAUAACCAUAUUCAGGGUGCUUCAGAACUCUAAGGCAAUUUGAAAAAAAAAAACCUGGCACUUUAAUUAUUGUCAUAGGUCCUGAGCAAACCUUGAAUU ----------..................((((((((((((.(((.((.(((.(((((((((((.................)))))..)))))).))))).))))))..))))))))). ( -24.23) >DroSim_CAF1 18836 112 - 1 -----GUUAUAUAUACAUAUAUAACCAUAUUCAGGGUGCUUCAGUUCUCUAAGGCAAUUUGAAAAA-AAAAAGCUGGCACUUUAAUUAUUGUCAUAGGUCCUGAGCAAACGUUGAAUU -----((((((((....))))))))...((((((.(((((.(((..(.(((.((((((((....))-).((((......)))).....))))).))))..))))))..)).)))))). ( -24.90) >DroEre_CAF1 15971 111 - 1 GAGUAUGUAUAUAUAUAUGUAUAACCAUAUUCAGGGUGCUUCAGAACUCUAAGGCAUUUUGAAA------AAGCUGGCACUCUAAUUAUUGCCAUAGGUCCAGAACAAGUCUUGAAU- (((((((..(((((....)))))..)))))))(((((((((.((....)).)))))))))((..------...((((.((.(((..........)))))))))......))......- ( -24.70) >consensus _______UAUAUAUACAUAUAUAACCAUAUUCAGGGUGCUUCAGAACUCUAAGGCAAUUUGAAAAA_AAAAAGCUGGCACUUUAAUUAUUGUCAUAGGUCCUGAGCAAACCUUGAAUU ............................(((((((((..(((((.((.(((.((((((.............................)))))).))))).)))))...))))))))). (-17.40 = -17.85 + 0.45)


| Location | 16,159,049 – 16,159,153 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -20.22 |
| Energy contribution | -22.00 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16159049 104 + 23771897 GCCAGGUUUUUUUUUUUCAAAUUGCCUUAGAGUUCUGAAGCACCCUGAAUAUGGUUAUAUAUGUAUAU--------------AUGUAUAUAUUGUUCAGCCAGACAUUCGAUUGCUGC ((.((((..(((......)))..))))..((((((((.......(((((((....(((((((((....--------------))))))))).))))))).)))).))))....))... ( -19.10) >DroSim_CAF1 18876 111 + 1 GCCAGCUUUUU-UUUUUCAAAUUGCCUUAGAGAACUGAAGCACCCUGAAUAUGGUUAUAUAUGUAUAUAUAAC------AAUAUAUACAUAUUCUUUAGCCAGACAUUCGAUUGCUGC ..((((.....-..........(((.((((....)))).)))...(((((.((((((.((((((((((((...------.))))))))))))....))))))...)))))...)))). ( -29.00) >DroEre_CAF1 16010 112 + 1 GCCAGCUU------UUUCAAAAUGCCUUAGAGUUCUGAAGCACCCUGAAUAUGGUUAUACAUAUAUAUAUACAUACUCGUAUAUGUACGUAUUAUUCAGCCAGACAUUCGAUUGCCGC ((..((..------..((....(((.((((....)))).)))....((((.(((((((((.(((((((((........))))))))).)))).....)))))...))))))..)).)) ( -22.40) >consensus GCCAGCUUUUU_UUUUUCAAAUUGCCUUAGAGUUCUGAAGCACCCUGAAUAUGGUUAUAUAUGUAUAUAUA_________AUAUGUACAUAUUAUUCAGCCAGACAUUCGAUUGCUGC ..((((................(((.((((....)))).)))...(((((.(((((..((((((((((((((......)))))))))))))).....)))))...)))))...)))). (-20.22 = -22.00 + 1.78)



| Location | 16,159,049 – 16,159,153 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -24.11 |
| Consensus MFE | -17.97 |
| Energy contribution | -19.20 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16159049 104 - 23771897 GCAGCAAUCGAAUGUCUGGCUGAACAAUAUAUACAU--------------AUAUACAUAUAUAACCAUAUUCAGGGUGCUUCAGAACUCUAAGGCAAUUUGAAAAAAAAAAACCUGGC .((((.............)))).............(--------------((((....))))).......(((((.(((((.((....)).))))).(((....))).....))))). ( -15.52) >DroSim_CAF1 18876 111 - 1 GCAGCAAUCGAAUGUCUGGCUAAAGAAUAUGUAUAUAUU------GUUAUAUAUACAUAUAUAACCAUAUUCAGGGUGCUUCAGUUCUCUAAGGCAAUUUGAAAAA-AAAAAGCUGGC .((((......((((((......)))((((((((((((.------...)))))))))))).....))).((((((.(((((.((....)).))))).))))))...-.....)))).. ( -28.00) >DroEre_CAF1 16010 112 - 1 GCGGCAAUCGAAUGUCUGGCUGAAUAAUACGUACAUAUACGAGUAUGUAUAUAUAUAUGUAUAACCAUAUUCAGGGUGCUUCAGAACUCUAAGGCAUUUUGAAA------AAGCUGGC ((((((......)))).((((.....(((((((.((((((......))))))...))))))).......((((((((((((.((....)).)))))))))))).------.)))).)) ( -28.80) >consensus GCAGCAAUCGAAUGUCUGGCUGAACAAUAUGUACAUAU_________UAUAUAUACAUAUAUAACCAUAUUCAGGGUGCUUCAGAACUCUAAGGCAAUUUGAAAAA_AAAAAGCUGGC .((((...........(((.((....(((((((.((((((......)))))).))))))).)).)))..(((((..(((((.((....)).)))))..))))).........)))).. (-17.97 = -19.20 + 1.23)


| Location | 16,159,087 – 16,159,193 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -26.24 |
| Energy contribution | -28.13 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16159087 106 + 23771897 AGCACCCUGAAUAUGGUUAUAUAUGUAUAU--------------AUGUAUAUAUUGUUCAGCCAGACAUUCGAUUGCUGCAGCCGGCAGCAGCAGCGGCAGCAAAACAAGGGCAAACAAU ....((((((((.(((((..((((((((..--------------..)))))))).....)))))...))))..(((((((.((..((....)).)).)))))))....))))........ ( -31.50) >DroSim_CAF1 18913 108 + 1 AGCACCCUGAAUAUGGUUAUAUAUGUAUAUAUAAC------AAUAUAUACAUAUUCUUUAGCCAGACAUUCGAUUGCUGCAGCCGGCA------GCGGCAGCAAAACAAGGGCAAACAAU ....((((((((.((((((.((((((((((((...------.))))))))))))....))))))...))))..(((((((.((.....------)).)))))))....))))........ ( -38.00) >DroEre_CAF1 16042 111 + 1 AGCACCCUGAAUAUGGUUAUACAUAUAUAUAUACAUACUCGUAUAUGUACGUAUUAUUCAGCCAGACAUUCGAUUGCCGCAGCCGGCA------G---CAGCAGAACAAGGGCAAACAAU ....((((((((.(((((((((.(((((((((........))))))))).)))).....)))))...))))(.((((((....)))))------)---).........))))........ ( -27.80) >consensus AGCACCCUGAAUAUGGUUAUAUAUGUAUAUAUA_________AUAUGUACAUAUUAUUCAGCCAGACAUUCGAUUGCUGCAGCCGGCA______GCGGCAGCAAAACAAGGGCAAACAAU ....((((((((.(((((..((((((((((((((......)))))))))))))).....)))))...))))..(((((((.((...........)).)))))))....))))........ (-26.24 = -28.13 + 1.90)



| Location | 16,159,087 – 16,159,193 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -20.74 |
| Energy contribution | -22.30 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16159087 106 - 23771897 AUUGUUUGCCCUUGUUUUGCUGCCGCUGCUGCUGCCGGCUGCAGCAAUCGAAUGUCUGGCUGAACAAUAUAUACAU--------------AUAUACAUAUAUAACCAUAUUCAGGGUGCU .......(((((....(((((((.(((((....).)))).)))))))..(((((..(((........(((((....--------------))))).........)))))))))))))... ( -28.13) >DroSim_CAF1 18913 108 - 1 AUUGUUUGCCCUUGUUUUGCUGCCGC------UGCCGGCUGCAGCAAUCGAAUGUCUGGCUAAAGAAUAUGUAUAUAUU------GUUAUAUAUACAUAUAUAACCAUAUUCAGGGUGCU .......(((((....(((((((.((------.....)).)))))))..((((((((......)))((((((((((((.------...)))))))))))).......))))))))))... ( -35.30) >DroEre_CAF1 16042 111 - 1 AUUGUUUGCCCUUGUUCUGCUG---C------UGCCGGCUGCGGCAAUCGAAUGUCUGGCUGAAUAAUACGUACAUAUACGAGUAUGUAUAUAUAUAUGUAUAACCAUAUUCAGGGUGCU ((((((((((..((((((((((---(------........))))))...)))))...))).)))))))..((((......(((((((..(((((....)))))..)))))))...)))). ( -30.10) >consensus AUUGUUUGCCCUUGUUUUGCUGCCGC______UGCCGGCUGCAGCAAUCGAAUGUCUGGCUGAACAAUAUGUACAUAU_________UAUAUAUACAUAUAUAACCAUAUUCAGGGUGCU .......(((((.....((((((.((...........)).))))))...(((((..(((.((....(((((((.((((((......)))))).))))))).)).)))))))))))))... (-20.74 = -22.30 + 1.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:35 2006