
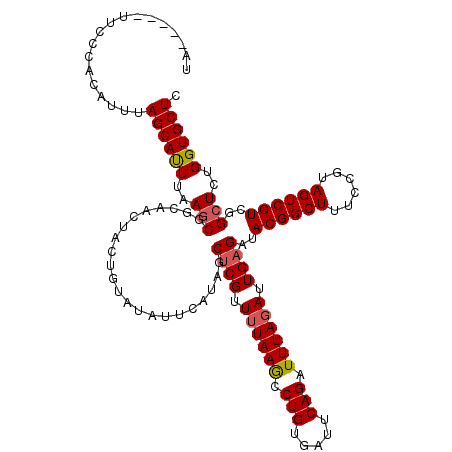
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,156,997 – 16,157,270 |
| Length | 273 |
| Max. P | 0.901966 |

| Location | 16,156,997 – 16,157,112 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.57 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16156997 115 + 23771897 UA-----UUCCUACAUUUAGCACUUAAGCGGCAACUACUGUAUAUUCAUAGCUCGUUUUAAGCCUGUGAUUCAGAUUUAGAUUGAGUUACGGCUUCCCGUAGUCGUCGGCUCUGGUGCUC ..-----...........((((((..((((((.(((((..........(((((((.(((((..(((.....)))..))))).))))))).((....))))))).))).)))..)))))). ( -35.00) >DroSec_CAF1 14513 115 + 1 UA-----UUCCCACAUUUAGCAUUUAAGCAGCAAAUACUGUAUAUUCAUAGCUCGAUUUAAACCUGUGAUUCAGAUUUAGAUUGAGAUACGGCUUUCCGUAGUCGUCGGCUCUGGUGCUC ..-----...........((((((..(((......................(((((((((((.(((.....))).)))))))))))..((((((......))))))..)))..)))))). ( -29.90) >DroSim_CAF1 15827 115 + 1 UA-----UUAUCACAUUUAGCAUUUAAGCGACCACUACUGUAUAUUCAUAGCUCGUUUUAAGCCUGUGAUUCAGAUUUAGAUUGAGAUACGGCUUUCCGUAGUCGUCGGCUGUGGUGCUC ..-----...........((((((..((((((.(((((.............((((.(((((..(((.....)))..))))).))))....((....))))))).))).)))..)))))). ( -28.40) >DroEre_CAF1 13883 120 + 1 UACGCACCUCCAAGAUUUAGCAUUUAAACCGCAACUACUGUAUAUUCAUGCCCCGUUGUAAGUCUGUGAUUCAGAUUUAGAUUGAGAUACGGCUUUCCAUAGUCGUCGGCUCUGGUGCUC ...(((((.....((((((...........(((((....((((....))))...))))).((((((.....))))))))))))(((..((((((......))))))...))).))))).. ( -30.20) >consensus UA_____UUCCCACAUUUAGCAUUUAAGCGGCAACUACUGUAUAUUCAUAGCUCGUUUUAAGCCUGUGAUUCAGAUUUAGAUUGAGAUACGGCUUUCCGUAGUCGUCGGCUCUGGUGCUC ..................((((((..(((......................((((.((((((.(((.....))).)))))).))))..((((((......))))))..)))..)))))). (-22.20 = -22.57 + 0.37)


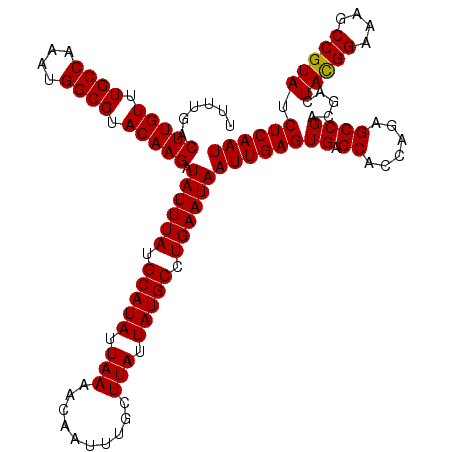
| Location | 16,156,997 – 16,157,112 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -29.39 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.34 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16156997 115 - 23771897 GAGCACCAGAGCCGACGACUACGGGAAGCCGUAACUCAAUCUAAAUCUGAAUCACAGGCUUAAAACGAGCUAUGAAUAUACAGUAGUUGCCGCUUAAGUGCUAAAUGUAGGAA-----UA .(((((..((((.(.(((((((((....))(((.(((....(((..(((.....)))..)))....))).))).........)))))))).))))..)))))...........-----.. ( -31.20) >DroSec_CAF1 14513 115 - 1 GAGCACCAGAGCCGACGACUACGGAAAGCCGUAUCUCAAUCUAAAUCUGAAUCACAGGUUUAAAUCGAGCUAUGAAUAUACAGUAUUUGCUGCUUAAAUGCUAAAUGUGGGAA-----UA .((((.((.((((((.((.(((((....)))))))......((((((((.....))))))))..))).))).))..((..((((....))))..))..))))...........-----.. ( -26.10) >DroSim_CAF1 15827 115 - 1 GAGCACCACAGCCGACGACUACGGAAAGCCGUAUCUCAAUCUAAAUCUGAAUCACAGGCUUAAAACGAGCUAUGAAUAUACAGUAGUGGUCGCUUAAAUGCUAAAUGUGAUAA-----UA .((((....(((.(((.(((((((....))((((.(((........(((.....)))((((.....))))..)))..)))).))))).))))))....))))...........-----.. ( -27.50) >DroEre_CAF1 13883 120 - 1 GAGCACCAGAGCCGACGACUAUGGAAAGCCGUAUCUCAAUCUAAAUCUGAAUCACAGACUUACAACGGGGCAUGAAUAUACAGUAGUUGCGGUUUAAAUGCUAAAUCUUGGAGGUGCGUA ..(((((.((((((.(((((((.......(((..(((....(((.((((.....)))).)))....)))..)))........))))))))))))).....(((.....))).)))))... ( -32.76) >consensus GAGCACCAGAGCCGACGACUACGGAAAGCCGUAUCUCAAUCUAAAUCUGAAUCACAGGCUUAAAACGAGCUAUGAAUAUACAGUAGUUGCCGCUUAAAUGCUAAAUGUGGGAA_____UA .((((...((((.(.(((((((.......((((.(((....(((.((((.....)))).)))....))).))))........)))))))).))))...)))).................. (-21.27 = -21.34 + 0.06)

| Location | 16,157,072 – 16,157,192 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -28.58 |
| Energy contribution | -28.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16157072 120 + 23771897 AUUGAGUUACGGCUUCCCGUAGUCGUCGGCUCUGGUGCUCACUCAAUUAUUCAGGCAUAAUAAGCAAAUUGUUUUAAUAUGCAUAAAUAUCUUGUACGCCAUUUGGCAAACAAGUCAAAA ...(((((((((((......)))))).)))))(((((..((......((((...(((((..((((.....))))...)))))...))))...))..)))))((((((......)))))). ( -30.20) >DroSec_CAF1 14588 120 + 1 AUUGAGAUACGGCUUUCCGUAGUCGUCGGCUCUGGUGCUCACUCAAUUAUUCAGGCAUAAUAAGCAAAUUGUUUUAAUAUGCAUAAAUAUCUUGUACGCCAUUUGGCAAACAAGUCAAAA ((((((..((((((......)))))).(((......)))..))))))((((...(((((..((((.....))))...)))))...)))).(((((..(((....)))..)))))...... ( -28.70) >DroSim_CAF1 15902 120 + 1 AUUGAGAUACGGCUUUCCGUAGUCGUCGGCUGUGGUGCUCACUCAAUUAUUCAGGCAUAAUAAGCAAAUUGUUUUAAUAUGCAUAAAUAUCUUGUACGCCAUUUGGCAAACAAGUCAAAA ((((((....(((...(((((((.....))))))).)))..))))))((((...(((((..((((.....))))...)))))...)))).(((((..(((....)))..)))))...... ( -29.40) >DroEre_CAF1 13963 120 + 1 AUUGAGAUACGGCUUUCCAUAGUCGUCGGCUCUGGUGCUCACUCAAUUAUUCAGGCAUAAUAAGCAAAUUGUUUUAAUAUGCAUAAAUAUCUUGUACGCCAUUUGGCAAACAAGUCAAAA ((((((....(((...(((.(((.....))).))).)))..))))))((((...(((((..((((.....))))...)))))...)))).(((((..(((....)))..)))))...... ( -28.70) >consensus AUUGAGAUACGGCUUUCCGUAGUCGUCGGCUCUGGUGCUCACUCAAUUAUUCAGGCAUAAUAAGCAAAUUGUUUUAAUAUGCAUAAAUAUCUUGUACGCCAUUUGGCAAACAAGUCAAAA ((((((..((((((......)))))).(((......)))..))))))((((...(((((..((((.....))))...)))))...)))).(((((..(((....)))..)))))...... (-28.58 = -28.57 + -0.00)



| Location | 16,157,072 – 16,157,192 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -30.29 |
| Energy contribution | -30.10 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16157072 120 - 23771897 UUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGCAUAUUAAAACAAUUUGCUUAUUAUGCCUGAAUAAUUGAGUGAGCACCAGAGCCGACGACUACGGGAAGCCGUAACUCAAU ......(((((.((((....)))).))))).((((((.(((((.(((..........))).))))).))))))((((((((.((......)))......(((((....)))))))))))) ( -33.60) >DroSec_CAF1 14588 120 - 1 UUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGCAUAUUAAAACAAUUUGCUUAUUAUGCCUGAAUAAUUGAGUGAGCACCAGAGCCGACGACUACGGAAAGCCGUAUCUCAAU ......(((((.((((....)))).))))).((((((.(((((.(((..........))).))))).))))))((((((((.((......)))).....(((((....))))).)))))) ( -30.00) >DroSim_CAF1 15902 120 - 1 UUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGCAUAUUAAAACAAUUUGCUUAUUAUGCCUGAAUAAUUGAGUGAGCACCACAGCCGACGACUACGGAAAGCCGUAUCUCAAU ......(((((.((((....)))).))))).((((((.(((((.(((..........))).))))).))))))((((((((.((......)))).....(((((....))))).)))))) ( -30.00) >DroEre_CAF1 13963 120 - 1 UUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGCAUAUUAAAACAAUUUGCUUAUUAUGCCUGAAUAAUUGAGUGAGCACCAGAGCCGACGACUAUGGAAAGCCGUAUCUCAAU ......(((((.((((....)))).))))).((((((.(((((.(((..........))).))))).))))))((((((((.((......)))).....(((((....))))).)))))) ( -28.20) >consensus UUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGCAUAUUAAAACAAUUUGCUUAUUAUGCCUGAAUAAUUGAGUGAGCACCAGAGCCGACGACUACGGAAAGCCGUAUCUCAAU ......(((((.((((....)))).))))).((((((.(((((.(((..........))).))))).))))))((((((((.((......)))).....(((((....))))).)))))) (-30.29 = -30.10 + -0.19)


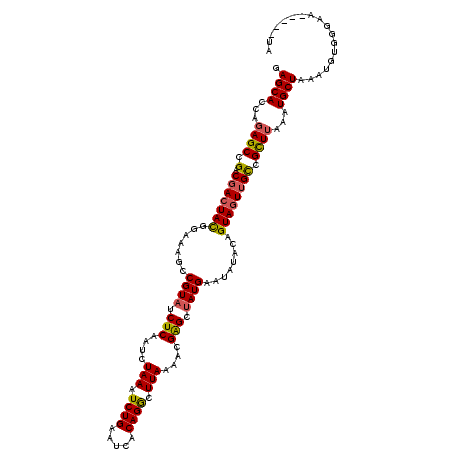
| Location | 16,157,112 – 16,157,232 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -34.00 |
| Energy contribution | -34.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.69 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16157112 120 + 23771897 ACUCAAUUAUUCAGGCAUAAUAAGCAAAUUGUUUUAAUAUGCAUAAAUAUCUUGUACGCCAUUUGGCAAACAAGUCAAAACUGCCAGACAAACUGGCAGGCAGUGGCGCAGACAGACAGC .......((((...(((((..((((.....))))...)))))...))))..((((.(((((((((((......)))))..(((((((.....)))))))...))))))...))))..... ( -34.00) >DroSec_CAF1 14628 120 + 1 ACUCAAUUAUUCAGGCAUAAUAAGCAAAUUGUUUUAAUAUGCAUAAAUAUCUUGUACGCCAUUUGGCAAACAAGUCAAAACUGCCAGACAAACUGGCAGGCAGUGGCGCAGACAGACAGC .......((((...(((((..((((.....))))...)))))...))))..((((.(((((((((((......)))))..(((((((.....)))))))...))))))...))))..... ( -34.00) >DroSim_CAF1 15942 120 + 1 ACUCAAUUAUUCAGGCAUAAUAAGCAAAUUGUUUUAAUAUGCAUAAAUAUCUUGUACGCCAUUUGGCAAACAAGUCAAAACUGCCAGACAAACUGGCAGGCAGUGGCGCAGACAGACAGC .......((((...(((((..((((.....))))...)))))...))))..((((.(((((((((((......)))))..(((((((.....)))))))...))))))...))))..... ( -34.00) >DroEre_CAF1 14003 120 + 1 ACUCAAUUAUUCAGGCAUAAUAAGCAAAUUGUUUUAAUAUGCAUAAAUAUCUUGUACGCCAUUUGGCAAACAAGUCAAAACUGCCAGACAAACUGGCAGGCAGUGGCGCAGACAGACAGC .......((((...(((((..((((.....))))...)))))...))))..((((.(((((((((((......)))))..(((((((.....)))))))...))))))...))))..... ( -34.00) >consensus ACUCAAUUAUUCAGGCAUAAUAAGCAAAUUGUUUUAAUAUGCAUAAAUAUCUUGUACGCCAUUUGGCAAACAAGUCAAAACUGCCAGACAAACUGGCAGGCAGUGGCGCAGACAGACAGC .......((((...(((((..((((.....))))...)))))...))))..((((.(((((((((((......)))))..(((((((.....)))))))...))))))...))))..... (-34.00 = -34.00 + 0.00)



| Location | 16,157,112 – 16,157,232 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16157112 120 - 23771897 GCUGUCUGUCUGCGCCACUGCCUGCCAGUUUGUCUGGCAGUUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGCAUAUUAAAACAAUUUGCUUAUUAUGCCUGAAUAAUUGAGU (((.(((...(((((((....(((((((.....))))))).((((.(......).)))))))))))..)))((((((.(((((.(((..........))).))))).))))))....))) ( -33.00) >DroSec_CAF1 14628 120 - 1 GCUGUCUGUCUGCGCCACUGCCUGCCAGUUUGUCUGGCAGUUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGCAUAUUAAAACAAUUUGCUUAUUAUGCCUGAAUAAUUGAGU (((.(((...(((((((....(((((((.....))))))).((((.(......).)))))))))))..)))((((((.(((((.(((..........))).))))).))))))....))) ( -33.00) >DroSim_CAF1 15942 120 - 1 GCUGUCUGUCUGCGCCACUGCCUGCCAGUUUGUCUGGCAGUUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGCAUAUUAAAACAAUUUGCUUAUUAUGCCUGAAUAAUUGAGU (((.(((...(((((((....(((((((.....))))))).((((.(......).)))))))))))..)))((((((.(((((.(((..........))).))))).))))))....))) ( -33.00) >DroEre_CAF1 14003 120 - 1 GCUGUCUGUCUGCGCCACUGCCUGCCAGUUUGUCUGGCAGUUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGCAUAUUAAAACAAUUUGCUUAUUAUGCCUGAAUAAUUGAGU (((.(((...(((((((....(((((((.....))))))).((((.(......).)))))))))))..)))((((((.(((((.(((..........))).))))).))))))....))) ( -33.00) >consensus GCUGUCUGUCUGCGCCACUGCCUGCCAGUUUGUCUGGCAGUUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGCAUAUUAAAACAAUUUGCUUAUUAUGCCUGAAUAAUUGAGU (((.(((...(((((((....(((((((.....))))))).((((.(......).)))))))))))..)))((((((.(((((.(((..........))).))))).))))))....))) (-33.00 = -33.00 + 0.00)


| Location | 16,157,152 – 16,157,270 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.59 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16157152 118 - 23771897 GUAACCCCCCUUUCCACAACACCCCCCC--CCCCCCUGCAGCUGUCUGUCUGCGCCACUGCCUGCCAGUUUGUCUGGCAGUUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGC ............................--.......(((..(((((...(((((((....(((((((.....))))))).((((.(......).)))))))))))..)))))....))) ( -29.20) >DroSec_CAF1 14668 117 - 1 GUAAGUCCCCUUUCCA-ACCAACCCCCU--CCCCCCUGCAGCUGUCUGUCUGCGCCACUGCCUGCCAGUUUGUCUGGCAGUUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGC ((((((..........-..(((......--.......(((((.....).))))........(((((((.....)))))))..))).(((((.((((....)))).)))))..)))))).. ( -29.70) >DroSim_CAF1 15982 119 - 1 GUAAGCCCCCUUUCGA-ACCAACCCCCUUCCCCCCCUGCAGCUGUCUGUCUGCGCCACUGCCUGCCAGUUUGUCUGGCAGUUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGC (((((.......((((-(...................(((((.....).))))........(((((((.....))))))).)))))(((((.((((....)))).)))))...))))).. ( -30.60) >DroEre_CAF1 14043 102 - 1 ----GGCCCCUUUCG------------C--CCCCCCUGCAGCUGUCUGUCUGCGCCACUGCCUGCCAGUUUGUCUGGCAGUUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGC ----(((.......)------------)--)......(((..(((((...(((((((....(((((((.....))))))).((((.(......).)))))))))))..)))))....))) ( -31.70) >consensus GUAAGCCCCCUUUCCA_ACCAACCCCCC__CCCCCCUGCAGCUGUCUGUCUGCGCCACUGCCUGCCAGUUUGUCUGGCAGUUUUGACUUGUUUGCCAAAUGGCGUACAAGAUAUUUAUGC .....................................(((..(((((...(((((((....(((((((.....))))))).((((.(......).)))))))))))..)))))....))) (-29.20 = -29.20 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:26 2006