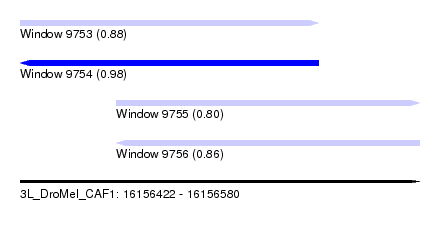
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,156,422 – 16,156,580 |
| Length | 158 |
| Max. P | 0.975752 |

| Location | 16,156,422 – 16,156,540 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -33.94 |
| Consensus MFE | -34.05 |
| Energy contribution | -33.86 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16156422 118 + 23771897 --UAUUUGCCCUGGUCCGUUGUCUCGCUUAACUUUUUUAUUGCCAGACGAAGCGCAAACAUUUGCGAGUAUUAAUUAACGCUUUGUUGAAGUUUGCGCUUCGAUGUCGCCGCCCAGACAU --........((((.(.((.((((.((..............)).))))(((((((((((.((.(((((..((....))..).)))).)).)))))))))))......)).).)))).... ( -33.54) >DroSec_CAF1 13808 120 + 1 UCUAUUUGCCCUGGUCCGUUGUCUCGCUUAACUUUUUUAUUGCCAGACGAAGCGCAAACAUUUGCGAGUAUUAAUUAACGCUUUGUUGAAGUUUGCGCUUCGAUGUCGCCGCCCAGACAU ..........((((.(.((.((((.((..............)).))))(((((((((((.((.(((((..((....))..).)))).)).)))))))))))......)).).)))).... ( -33.54) >DroSim_CAF1 15130 120 + 1 UCUAUUUGCCCUGGUCCGUUGUCUCGCUUAACUUUUUUAUUGCCAGACGAAGCGCAAACAUUUGCGAGUAUUAAUUAACGCUUUGUUGAAGUUUGCGCUUCGAUGUCGCCGCCCAGACAU ..........((((.(.((.((((.((..............)).))))(((((((((((.((.(((((..((....))..).)))).)).)))))))))))......)).).)))).... ( -33.54) >DroEre_CAF1 13384 120 + 1 UGUAUUUGCUCCGGUCCGUUGUCUCGCUUAACUUUUUUAUUGCCAGACGAAGCGCAAACAUUUGCGAGUAUUAAUUAACGCUUUGUUGAAGUUUGCGCUUCGAUGUCGCCGCCCAGACAU ....((((...((((.((((((((.((..............)).))))(((((((((((.((.(((((..((....))..).)))).)).)))))))))))))))..))))..))))... ( -35.14) >consensus UCUAUUUGCCCUGGUCCGUUGUCUCGCUUAACUUUUUUAUUGCCAGACGAAGCGCAAACAUUUGCGAGUAUUAAUUAACGCUUUGUUGAAGUUUGCGCUUCGAUGUCGCCGCCCAGACAU ....((((...((((.((((((((.((..............)).))))(((((((((((.((.(((((..((....))..).)))).)).)))))))))))))))..))))..))))... (-34.05 = -33.86 + -0.19)

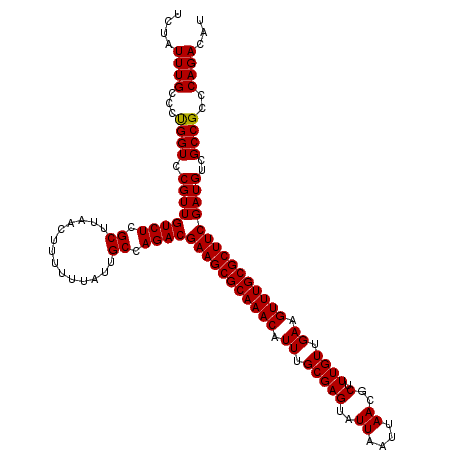
| Location | 16,156,422 – 16,156,540 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -36.01 |
| Energy contribution | -35.63 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.74 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16156422 118 - 23771897 AUGUCUGGGCGGCGACAUCGAAGCGCAAACUUCAACAAAGCGUUAAUUAAUACUCGCAAAUGUUUGCGCUUCGUCUGGCAAUAAAAAAGUUAAGCGAGACAACGGACCAGGGCAAAUA-- .(((((((.((........(((((((((((.........(((............)))....)))))))))))((((.((..............)).))))..))..))).))))....-- ( -35.66) >DroSec_CAF1 13808 120 - 1 AUGUCUGGGCGGCGACAUCGAAGCGCAAACUUCAACAAAGCGUUAAUUAAUACUCGCAAAUGUUUGCGCUUCGUCUGGCAAUAAAAAAGUUAAGCGAGACAACGGACCAGGGCAAAUAGA .(((((((.((........(((((((((((.........(((............)))....)))))))))))((((.((..............)).))))..))..))).))))...... ( -35.66) >DroSim_CAF1 15130 120 - 1 AUGUCUGGGCGGCGACAUCGAAGCGCAAACUUCAACAAAGCGUUAAUUAAUACUCGCAAAUGUUUGCGCUUCGUCUGGCAAUAAAAAAGUUAAGCGAGACAACGGACCAGGGCAAAUAGA .(((((((.((........(((((((((((.........(((............)))....)))))))))))((((.((..............)).))))..))..))).))))...... ( -35.66) >DroEre_CAF1 13384 120 - 1 AUGUCUGGGCGGCGACAUCGAAGCGCAAACUUCAACAAAGCGUUAAUUAAUACUCGCAAAUGUUUGCGCUUCGUCUGGCAAUAAAAAAGUUAAGCGAGACAACGGACCGGAGCAAAUACA .(((((((.((........(((((((((((.........(((............)))....)))))))))))((((.((..............)).))))..))..))))).))...... ( -35.56) >consensus AUGUCUGGGCGGCGACAUCGAAGCGCAAACUUCAACAAAGCGUUAAUUAAUACUCGCAAAUGUUUGCGCUUCGUCUGGCAAUAAAAAAGUUAAGCGAGACAACGGACCAGGGCAAAUAGA .(((((((.((........(((((((((((.........(((............)))....)))))))))))((((.((..............)).))))..))..))))).))...... (-36.01 = -35.63 + -0.37)

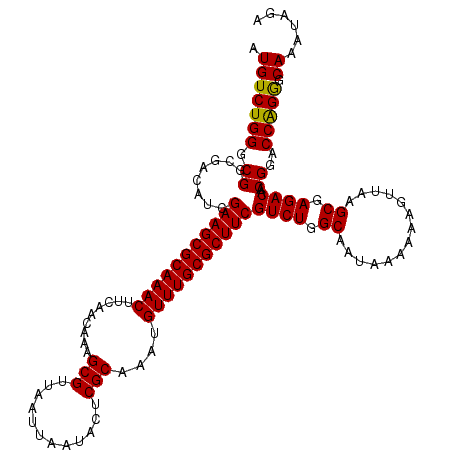
| Location | 16,156,460 – 16,156,580 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -38.17 |
| Energy contribution | -38.42 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16156460 120 + 23771897 UGCCAGACGAAGCGCAAACAUUUGCGAGUAUUAAUUAACGCUUUGUUGAAGUUUGCGCUUCGAUGUCGCCGCCCAGACAUCGAUGUAGACCUAGACGUUUGACGUAGACUUUGUCAAAUG ........(((((((((((.((.(((((..((....))..).)))).)).)))))))))))((((((........))))))..............(((((((((.......))))))))) ( -39.00) >DroSec_CAF1 13848 120 + 1 UGCCAGACGAAGCGCAAACAUUUGCGAGUAUUAAUUAACGCUUUGUUGAAGUUUGCGCUUCGAUGUCGCCGCCCAGACAUCGAUGUAGACCUAGACGUUUGACGUAGACUUUGUCAAAUG ........(((((((((((.((.(((((..((....))..).)))).)).)))))))))))((((((........))))))..............(((((((((.......))))))))) ( -39.00) >DroSim_CAF1 15170 120 + 1 UGCCAGACGAAGCGCAAACAUUUGCGAGUAUUAAUUAACGCUUUGUUGAAGUUUGCGCUUCGAUGUCGCCGCCCAGACAUCGAUGUAGACCUAGACGUUUGACGUAGACUUUGUCAAAUG ........(((((((((((.((.(((((..((....))..).)))).)).)))))))))))((((((........))))))..............(((((((((.......))))))))) ( -39.00) >DroEre_CAF1 13424 120 + 1 UGCCAGACGAAGCGCAAACAUUUGCGAGUAUUAAUUAACGCUUUGUUGAAGUUUGCGCUUCGAUGUCGCCGCCCAGACAUCGAUGUAGACCUAGACGCUUGACGUAGACUUUGUCAAAUG .((.....(((((((((((.((.(((((..((....))..).)))).)).)))))))))))((((((........))))))...............))((((((.......))))))... ( -36.30) >consensus UGCCAGACGAAGCGCAAACAUUUGCGAGUAUUAAUUAACGCUUUGUUGAAGUUUGCGCUUCGAUGUCGCCGCCCAGACAUCGAUGUAGACCUAGACGUUUGACGUAGACUUUGUCAAAUG ........(((((((((((.((.(((((..((....))..).)))).)).)))))))))))((((((........))))))..............(((((((((.......))))))))) (-38.17 = -38.42 + 0.25)


| Location | 16,156,460 – 16,156,580 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -36.57 |
| Consensus MFE | -36.76 |
| Energy contribution | -36.57 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16156460 120 - 23771897 CAUUUGACAAAGUCUACGUCAAACGUCUAGGUCUACAUCGAUGUCUGGGCGGCGACAUCGAAGCGCAAACUUCAACAAAGCGUUAAUUAAUACUCGCAAAUGUUUGCGCUUCGUCUGGCA ..((((((.........))))))((((((((((......)))..)))))))((.(...((((((((((((.........(((............)))....))))))))))))..).)). ( -36.72) >DroSec_CAF1 13848 120 - 1 CAUUUGACAAAGUCUACGUCAAACGUCUAGGUCUACAUCGAUGUCUGGGCGGCGACAUCGAAGCGCAAACUUCAACAAAGCGUUAAUUAAUACUCGCAAAUGUUUGCGCUUCGUCUGGCA ..((((((.........))))))((((((((((......)))..)))))))((.(...((((((((((((.........(((............)))....))))))))))))..).)). ( -36.72) >DroSim_CAF1 15170 120 - 1 CAUUUGACAAAGUCUACGUCAAACGUCUAGGUCUACAUCGAUGUCUGGGCGGCGACAUCGAAGCGCAAACUUCAACAAAGCGUUAAUUAAUACUCGCAAAUGUUUGCGCUUCGUCUGGCA ..((((((.........))))))((((((((((......)))..)))))))((.(...((((((((((((.........(((............)))....))))))))))))..).)). ( -36.72) >DroEre_CAF1 13424 120 - 1 CAUUUGACAAAGUCUACGUCAAGCGUCUAGGUCUACAUCGAUGUCUGGGCGGCGACAUCGAAGCGCAAACUUCAACAAAGCGUUAAUUAAUACUCGCAAAUGUUUGCGCUUCGUCUGGCA ..((((((.........))))))((((((((((......)))..)))))))((.(...((((((((((((.........(((............)))....))))))))))))..).)). ( -36.12) >consensus CAUUUGACAAAGUCUACGUCAAACGUCUAGGUCUACAUCGAUGUCUGGGCGGCGACAUCGAAGCGCAAACUUCAACAAAGCGUUAAUUAAUACUCGCAAAUGUUUGCGCUUCGUCUGGCA ..((((((.........))))))((((((((((......)))..)))))))((.(...((((((((((((.........(((............)))....))))))))))))..).)). (-36.76 = -36.57 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:20 2006