
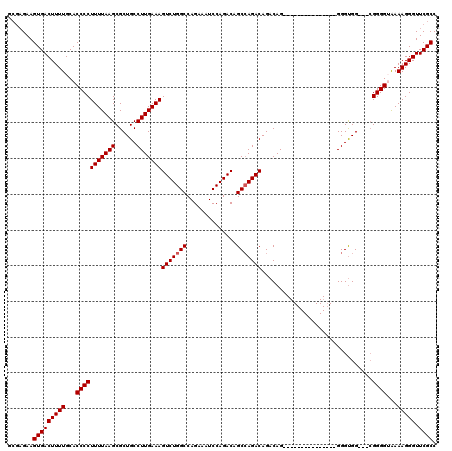
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,148,856 – 16,148,997 |
| Length | 141 |
| Max. P | 0.902160 |

| Location | 16,148,856 – 16,148,957 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.34 |
| Mean single sequence MFE | -42.59 |
| Consensus MFE | -27.67 |
| Energy contribution | -27.91 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16148856 101 + 23771897 ----GAAGUGACUUUUGCACCCCUUUUAAGCGCUGCCUUGAAAGUCUGGCCAGAAAUCCAGACAGCCAGACAGACAG---------------GGGUGGGAGGGGGGUGAAAGGGUUCGCC ----...(((((((((.((((((((((...(((..(((((...(((((((..............)))))))...)))---------------))))).)))))))))).))))).)))). ( -41.94) >DroSec_CAF1 10992 102 + 1 GCGAGAAGUGACUUUUGCACCCCUUUUAAGCGCUGCCUUGAAAGUCUGGCCAGAAAUCCAGACAGCCAGACAGACAG---------------GGGUGG---CGGGGAAAAAGGGUUCGCC (((((..(((.(....))))(((((((...(((..((((....(((((((..............))))))).....)---------------)))..)---))....)))))))))))). ( -38.94) >DroSim_CAF1 12373 102 + 1 GCGAGAAGUGACUUUUGCACCCCUUUUAAGCGCUGCCUUGAAAGUCUGGCCAGCAAUCCAGACAGCCAGACAGACAG---------------GGGUGG---CGGGGUAAAAGGGUUCGCC (((((..(((.(....))))((((((((..(((..((((....(((((((...(......)...))))))).....)---------------)))..)---))...))))))))))))). ( -42.80) >DroEre_CAF1 10693 116 + 1 CCGAGAAGUGACUUUUGCACCCCUUUUAAGCGCUGCCUUGAAAGUCUGGCCAGAAAUCCAGACAGCGAGACAGCCAGACAGCCAGACUGACAGGGCGG---CGGGG-AAAAGGGUUCGCC .((((..(((.(....))))(((((((...((((((((((..((((((((..(.....).....((......))......))))))))..))))))))---))...-))))))))))).. ( -46.70) >consensus GCGAGAAGUGACUUUUGCACCCCUUUUAAGCGCUGCCUUGAAAGUCUGGCCAGAAAUCCAGACAGCCAGACAGACAG_______________GGGUGG___CGGGGUAAAAGGGUUCGCC .......(((((((((...(((((((((((......)))))))(((((((..............)))))))...............................))))...))))).)))). (-27.67 = -27.91 + 0.25)


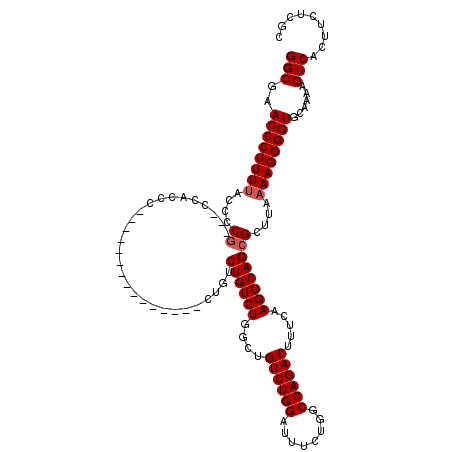
| Location | 16,148,856 – 16,148,957 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.34 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -24.28 |
| Energy contribution | -24.78 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16148856 101 - 23771897 GGCGAACCCUUUCACCCCCCUCCCACCC---------------CUGUCUGUCUGGCUGUCUGGAUUUCUGGCCAGACUUUCAAGGCAGCGCUUAAAAGGGGUGCAAAAGUCACUUC---- ((((((....)))..........(((((---------------((((((((((((..((((((........))))))..)).)))))).)).....))))))).....))).....---- ( -32.60) >DroSec_CAF1 10992 102 - 1 GGCGAACCCUUUUUCCCCG---CCACCC---------------CUGUCUGUCUGGCUGUCUGGAUUUCUGGCCAGACUUUCAAGGCAGCGCUUAAAAGGGGUGCAAAAGUCACUUCUCGC .((((.(((((((....((---(...((---------------.((...((((((((............))))))))...)).))..)))...)))))))((((....).)))...)))) ( -34.30) >DroSim_CAF1 12373 102 - 1 GGCGAACCCUUUUACCCCG---CCACCC---------------CUGUCUGUCUGGCUGUCUGGAUUGCUGGCCAGACUUUCAAGGCAGCGCUUAAAAGGGGUGCAAAAGUCACUUCUCGC .((((.((((((((...((---(...((---------------.((...(((((((((.(......).)))))))))...)).))..)))..))))))))((((....).)))...)))) ( -36.50) >DroEre_CAF1 10693 116 - 1 GGCGAACCCUUUU-CCCCG---CCGCCCUGUCAGUCUGGCUGUCUGGCUGUCUCGCUGUCUGGAUUUCUGGCCAGACUUUCAAGGCAGCGCUUAAAAGGGGUGCAAAAGUCACUUCUCGG ..(((.(((((((-...((---(.(((.((..((((((((((((..((.(.....).).)..)))....)))))))))..)).))).)))...)))))))((((....).)))...))). ( -44.50) >consensus GGCGAACCCUUUUACCCCG___CCACCC_______________CUGUCUGUCUGGCUGUCUGGAUUUCUGGCCAGACUUUCAAGGCAGCGCUUAAAAGGGGUGCAAAAGUCACUUCUCGC (((..((((((((....((............................((((((....((((((........)))))).....))))))))....))))))))......)))......... (-24.28 = -24.78 + 0.50)

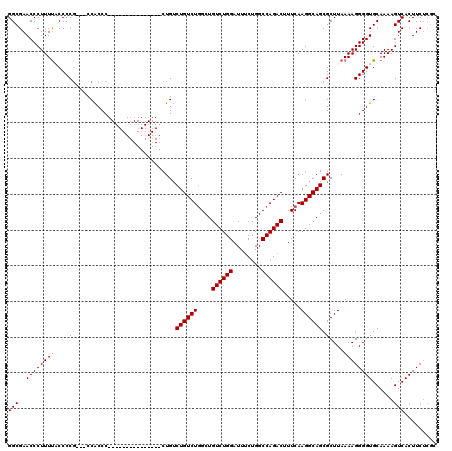
| Location | 16,148,892 – 16,148,997 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.09 |
| Mean single sequence MFE | -46.78 |
| Consensus MFE | -29.77 |
| Energy contribution | -29.64 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16148892 105 + 23771897 AAAGUCUGGCCAGAAAUCCAGACAGCCAGACAGACAG---------------GGGUGGGAGGGGGGUGAAAGGGUUCGCCCCCGCUGCGCCCCUGUCCAUCAUUUCGAUUGAGAAAUACU ...(((((((..............))))))).(((((---------------(((((.(..(((((((((....)))))))))..).))))))))))....(((((......)))))... ( -52.24) >DroSec_CAF1 11032 102 + 1 AAAGUCUGGCCAGAAAUCCAGACAGCCAGACAGACAG---------------GGGUGG---CGGGGAAAAAGGGUUCGCCCCCGCUGCGCCCCUGUCCAUCGUUUCGAUUGAGAAAUACU ...(((((((..............))))))).(((((---------------((((((---(((((.............))))))..))))))))))....(((((......)))))... ( -45.66) >DroSim_CAF1 12413 102 + 1 AAAGUCUGGCCAGCAAUCCAGACAGCCAGACAGACAG---------------GGGUGG---CGGGGUAAAAGGGUUCGCCCCCGCUGCGCCCCUGUCCAUCAUUUCGAUUGAGAAAUACU ...(((((((...(......)...))))))).(((((---------------((((((---(((((.............))))))..))))))))))....(((((......)))))... ( -45.92) >DroEre_CAF1 10733 116 + 1 AAAGUCUGGCCAGAAAUCCAGACAGCGAGACAGCCAGACAGCCAGACUGACAGGGCGG---CGGGG-AAAAGGGUUCGCCCCCGCUGCGCCCCUGUCCACGAUUUCGAUUGAGAAAUACU ..((((((((..(.....).....((......))......))))))))((((((((((---(((((-............)))))))))...))))))....(((((......)))))... ( -43.30) >consensus AAAGUCUGGCCAGAAAUCCAGACAGCCAGACAGACAG_______________GGGUGG___CGGGGUAAAAGGGUUCGCCCCCGCUGCGCCCCUGUCCAUCAUUUCGAUUGAGAAAUACU ...((((((........)))))).....(((((...................(((((....(((((.............)))))...))))))))))....(((((......)))))... (-29.77 = -29.64 + -0.13)



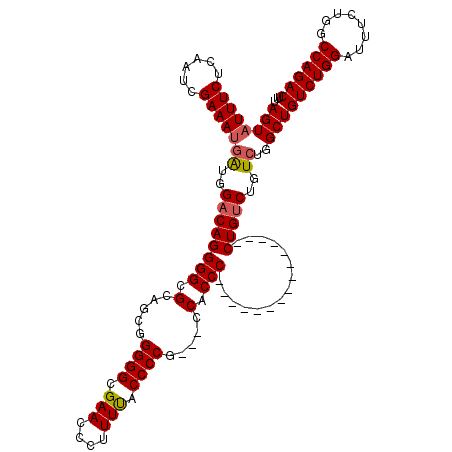
| Location | 16,148,892 – 16,148,997 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.09 |
| Mean single sequence MFE | -46.98 |
| Consensus MFE | -29.06 |
| Energy contribution | -29.44 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16148892 105 - 23771897 AGUAUUUCUCAAUCGAAAUGAUGGACAGGGGCGCAGCGGGGGCGAACCCUUUCACCCCCCUCCCACCC---------------CUGUCUGUCUGGCUGUCUGGAUUUCUGGCCAGACUUU ((((((((......)))))((((((((((((.(.((.(((((.(((....))).)))))))..).)))---------------)))))))))..)))((((((........))))))... ( -47.20) >DroSec_CAF1 11032 102 - 1 AGUAUUUCUCAAUCGAAACGAUGGACAGGGGCGCAGCGGGGGCGAACCCUUUUUCCCCG---CCACCC---------------CUGUCUGUCUGGCUGUCUGGAUUUCUGGCCAGACUUU (((.((((......)))).((((((((((((.(..(((((((.((.....)).))))))---)).)))---------------)))))))))..)))((((((........))))))... ( -45.20) >DroSim_CAF1 12413 102 - 1 AGUAUUUCUCAAUCGAAAUGAUGGACAGGGGCGCAGCGGGGGCGAACCCUUUUACCCCG---CCACCC---------------CUGUCUGUCUGGCUGUCUGGAUUGCUGGCCAGACUUU ..((((((......))))))..(((((((((.(..((((((.............)))))---)).)))---------------))))))(((((((((.(......).)))))))))... ( -46.92) >DroEre_CAF1 10733 116 - 1 AGUAUUUCUCAAUCGAAAUCGUGGACAGGGGCGCAGCGGGGGCGAACCCUUUU-CCCCG---CCGCCCUGUCAGUCUGGCUGUCUGGCUGUCUCGCUGUCUGGAUUUCUGGCCAGACUUU .(.(((((......))))))(((((((((((((..(((((((.((....)).)-)))))---)))))))(((((.(.....).))))))))).))).((((((........))))))... ( -48.60) >consensus AGUAUUUCUCAAUCGAAAUGAUGGACAGGGGCGCAGCGGGGGCGAACCCUUUUACCCCG___CCACCC_______________CUGUCUGUCUGGCUGUCUGGAUUUCUGGCCAGACUUU ((((((((......)))))((..((((((((.(.....((((.(((....))).)))).....).)))...............)))))..))..)))((((((........))))))... (-29.06 = -29.44 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:12 2006