| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
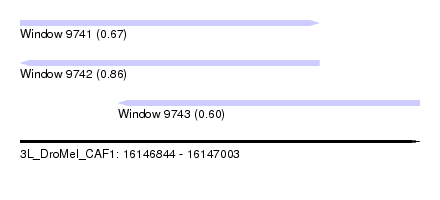
| Location | 16,146,844 – 16,147,003 |
| Length | 159 |
| Max. P | 0.864045 |

| Location | 16,146,844 – 16,146,963 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -25.36 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16146844 119 + 23771897 UUUAAAUU-UUAAACGAAUCCAAUAUCUAAAAUGAUUUCUCUCAGUGCACCCAGCAUAAGAGACACUCGUCGCAUGUGCAACGCAUGCCAUCUCGCUCUCGCCCACAGCUGCGUAUGCUG ........-....((((.......(((......))).(((((..((((.....)))).)))))...)))).(((((((...)))))))......((...(((........)))...)).. ( -24.30) >DroSec_CAF1 8868 114 + 1 -----GCU-UAAGAUAAAUCAAAUAUUUAAAGUGAUUUCUCUCAGUGCAUCCAGCAUAAGAGACACUCGUCGCAUGUGCAACGCAUGCCAUCUCGCUCUCGCCCGCAGCUGCGUGUGCUG -----...-..(((.((((((...........)))))).)))(((..(((.((((...((((((....)))(((((((...)))))))..))).((........)).)))).)))..))) ( -32.30) >DroSim_CAF1 9624 114 + 1 -----GCU-UAAGAUAAAUCAAAUAUCUAAAGUGAUUUCUUUCAGUGCAUCCAGCAUAAGAGACACUCGUCGCAUGUGCAACGCAUGCCAUCUCGCUCUCGCCCACAGCUGCGUGUGCUG -----(((-(.(((((.......))))).)))).........(((..(((.((((...((((((....)))(((((((...)))))))..))).((....)).....)))).)))..))) ( -32.90) >DroEre_CAF1 8415 115 + 1 -----GUUCUUAGACUAGUCCUAUACUCAAAGUGCUUUCUCUCAGUGCAUCCAGCAUAAGAGACACUCGUCGCAUGUGCAACGCAUGCCAUCUCGCUCUCGCCCAUUGCUGCGAGUGCUG -----.......(((.(((....(((.....)))...(((((..((((.....)))).))))).))).)))(((((((...)))))))......((.(((((........))))).)).. ( -31.70) >consensus _____GCU_UAAGACAAAUCAAAUAUCUAAAGUGAUUUCUCUCAGUGCAUCCAGCAUAAGAGACACUCGUCGCAUGUGCAACGCAUGCCAUCUCGCUCUCGCCCACAGCUGCGUGUGCUG ..........................................((((((((.((((...((((((....)))(((((((...)))))))..))).((....)).....)))).)))))))) (-25.36 = -25.68 + 0.31)



| Location | 16,146,844 – 16,146,963 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -29.62 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16146844 119 - 23771897 CAGCAUACGCAGCUGUGGGCGAGAGCGAGAUGGCAUGCGUUGCACAUGCGACGAGUGUCUCUUAUGCUGGGUGCACUGAGAGAAAUCAUUUUAGAUAUUGGAUUCGUUUAA-AAUUUAAA (((..(((.((((.....((....))((((.(((((.((((((....)))))).)))))))))..)))).)))..))).((....))..(((((((.(((((....)))))-.))))))) ( -39.00) >DroSec_CAF1 8868 114 - 1 CAGCACACGCAGCUGCGGGCGAGAGCGAGAUGGCAUGCGUUGCACAUGCGACGAGUGUCUCUUAUGCUGGAUGCACUGAGAGAAAUCACUUUAAAUAUUUGAUUUAUCUUA-AGC----- (((((..(((....))).((....))((((.(((((.((((((....)))))).))))))))).))))).......(((((.((((((..(.....)..)))))).)))))-...----- ( -38.40) >DroSim_CAF1 9624 114 - 1 CAGCACACGCAGCUGUGGGCGAGAGCGAGAUGGCAUGCGUUGCACAUGCGACGAGUGUCUCUUAUGCUGGAUGCACUGAAAGAAAUCACUUUAGAUAUUUGAUUUAUCUUA-AGC----- (((..((..((((.....((....))((((.(((((.((((((....)))))).)))))))))..))))..))..)))..........(((.(((((.......))))).)-)).----- ( -34.90) >DroEre_CAF1 8415 115 - 1 CAGCACUCGCAGCAAUGGGCGAGAGCGAGAUGGCAUGCGUUGCACAUGCGACGAGUGUCUCUUAUGCUGGAUGCACUGAGAGAAAGCACUUUGAGUAUAGGACUAGUCUAAGAAC----- ..((.(((((........))))).)).....(((((.((((((....)))))).)))))(((((.(((((.((.(((.((((......)))).))).))...))))).)))))..----- ( -40.00) >consensus CAGCACACGCAGCUGUGGGCGAGAGCGAGAUGGCAUGCGUUGCACAUGCGACGAGUGUCUCUUAUGCUGGAUGCACUGAGAGAAAUCACUUUAAAUAUUGGAUUUAUCUAA_AAC_____ ..(((....((((.....((....))((((.(((((.((((((....)))))).)))))))))..))))..))).............................................. (-29.62 = -29.62 + 0.00)


| Location | 16,146,883 – 16,147,003 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -39.89 |
| Energy contribution | -39.70 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16146883 120 - 23771897 AAAGUCAUAAACUUUAUUUUCUGUUGCCUGCACAAAGGCGCAGCAUACGCAGCUGUGGGCGAGAGCGAGAUGGCAUGCGUUGCACAUGCGACGAGUGUCUCUUAUGCUGGGUGCACUGAG (((((.....)))))..........((((......)))).(((..(((.((((.....((....))((((.(((((.((((((....)))))).)))))))))..)))).)))..))).. ( -42.00) >DroSec_CAF1 8902 120 - 1 AAAGUCAUAAACUUUAUUUUCUGUUGCCUGCACAAAGGCGCAGCACACGCAGCUGCGGGCGAGAGCGAGAUGGCAUGCGUUGCACAUGCGACGAGUGUCUCUUAUGCUGGAUGCACUGAG (((((.....))))).....((.((((((((......))(((((.......))))))))))).)).((((.(((((.((((((....)))))).))))))))).(((.....)))..... ( -41.10) >DroSim_CAF1 9658 120 - 1 AAAGUCAUAAACUUUAUUUUCUGUUGCCUGCACAAAGGCGCAGCACACGCAGCUGUGGGCGAGAGCGAGAUGGCAUGCGUUGCACAUGCGACGAGUGUCUCUUAUGCUGGAUGCACUGAA ..((((((............((((.((((......))))))))......((((.....((....))((((.(((((.((((((....)))))).)))))))))..)))).))).)))... ( -39.50) >DroEre_CAF1 8450 120 - 1 GAAGUCAUAAACUUUAUUUUCUGUUGCCUGCAUAAAGGCGCAGCACUCGCAGCAAUGGGCGAGAGCGAGAUGGCAUGCGUUGCACAUGCGACGAGUGUCUCUUAUGCUGGAUGCACUGAG (((((.....)))))......(((.((((......)))))))(((.((.(((((....((....))((((.(((((.((((((....)))))).))))))))).))))))))))...... ( -42.60) >consensus AAAGUCAUAAACUUUAUUUUCUGUUGCCUGCACAAAGGCGCAGCACACGCAGCUGUGGGCGAGAGCGAGAUGGCAUGCGUUGCACAUGCGACGAGUGUCUCUUAUGCUGGAUGCACUGAG ..((((((............((((.((((......))))))))......((((.....((....))((((.(((((.((((((....)))))).)))))))))..)))).))).)))... (-39.89 = -39.70 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:08 2006