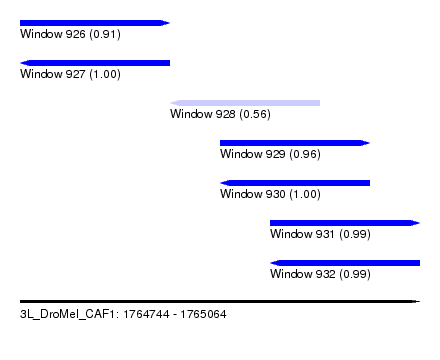
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,764,744 – 1,765,064 |
| Length | 320 |
| Max. P | 0.999990 |

| Location | 1,764,744 – 1,764,864 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -37.85 |
| Consensus MFE | -33.01 |
| Energy contribution | -33.03 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1764744 120 + 23771897 UGAUCUUUUGCAGAUUACGCCAGGUCUCCUCAAGAGCUUCCAUGGUGAACCACGUGUACGGGUUGGGUCCAACAUUAAAGCUCUUGAUCUUCUGGUCGAGUGCUGCCAAUGCCUUGAAGU ((((((.....))))))..(((((.....((((((((((((.(((....))).(....)))((((....))))....))))))))))...)))))(((((.((.......)))))))... ( -36.70) >DroVir_CAF1 1903 120 + 1 UGAUCUUUUGCAGAUUACGCCAGGUCUCCUCAAGAGCUUCCAUGGUGAACCAGGUGUACGGAUUUGGUCCAACAUUAAAGCUCUUGAUCUUCUGGUCGAGCGCUGCCAAUGCCUUAAAGU .........((((....(((((((.....((((((((((..(((.((.(((((((......))))))).)).)))..))))))))))...))))).))....)))).............. ( -38.80) >DroGri_CAF1 2395 120 + 1 UGAUCUUUUGCAAGUUACGCCAGGUCUCCUCAAGAGCUUCCAUGGUGAACCAUGUGUAUGGAUUGGGUCCAACAUUAAAGCUCUUGAUCUUCUGGUCGAGUGCUGCCAAUGCCUUAAAGU .........(((.((..(((((((.....((((((((((.(((((....)))))(((.(((((...))))))))...))))))))))...))))).))...))))).............. ( -37.50) >DroWil_CAF1 2671 120 + 1 UGAUCUUUUGCAGAUUACGCCAGGUCUCCUCAAGAGCUUCCAUGGUGAACCAUGUGUAUGGAUUCGGUCCAACAUUAAAGCUCUUGAUCUUCUGGUCGAGUGCUGCCAAUGCCUUAAAGU .........(((((((.(((((((.....((((((((((.(((((....)))))(((.(((((...))))))))...))))))))))...))))).))))).)))).............. ( -40.30) >DroMoj_CAF1 2240 120 + 1 UGAUCUUUUGCAGAUUACGCCAGGUCUCCUCAAGAGCUUCCAUGGUGAACCAGGUGUACGGAUUCGGUCCAACAUUAAAGCUCUUGAUCUUCUGGUCGAGUGCUGCCAAUGCCUUAAAGU .........(((((((.(((((((.....((((((((((...(((....)))(((((..((((...)))).))))).))))))))))...))))).))))).)))).............. ( -36.70) >DroAna_CAF1 2024 120 + 1 UGAUCUUUUGCAGGUUACGCCAGGUCUCCUCAAGAGCUUCCAUGGUGAACCAGGUGUAGGGAUUGGGUCCAACAUUAAAGCUCUUGAUCUUCUGGUCGAGUGCUGCCAAUGCCUUGAAAU ............((((.(((((((.(((.....)))...)).)))))))))((((((.((..(((....)))......(((.(((((((....))))))).))).)).))))))...... ( -37.10) >consensus UGAUCUUUUGCAGAUUACGCCAGGUCUCCUCAAGAGCUUCCAUGGUGAACCAGGUGUACGGAUUGGGUCCAACAUUAAAGCUCUUGAUCUUCUGGUCGAGUGCUGCCAAUGCCUUAAAGU .........((((....(((((((.....((((((((((..(((.((.(((..((......))..))).)).)))..))))))))))...))))).))....)))).............. (-33.01 = -33.03 + 0.03)

| Location | 1,764,744 – 1,764,864 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -40.89 |
| Consensus MFE | -39.88 |
| Energy contribution | -39.74 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.58 |
| SVM RNA-class probability | 0.999990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1764744 120 - 23771897 ACUUCAAGGCAUUGGCAGCACUCGACCAGAAGAUCAAGAGCUUUAAUGUUGGACCCAACCCGUACACGUGGUUCACCAUGGAAGCUCUUGAGGAGACCUGGCGUAAUCUGCAAAAGAUCA ..............((((....((.((((....(((((((((((...((((....)))).......(((((....))))))))))))))))(....)))))))....))))......... ( -40.70) >DroVir_CAF1 1903 120 - 1 ACUUUAAGGCAUUGGCAGCGCUCGACCAGAAGAUCAAGAGCUUUAAUGUUGGACCAAAUCCGUACACCUGGUUCACCAUGGAAGCUCUUGAGGAGACCUGGCGUAAUCUGCAAAAGAUCA ..............((((....((.((((....(((((((((((.(((.(((((((............))))))).))).)))))))))))(....)))))))....))))......... ( -41.80) >DroGri_CAF1 2395 120 - 1 ACUUUAAGGCAUUGGCAGCACUCGACCAGAAGAUCAAGAGCUUUAAUGUUGGACCCAAUCCAUACACAUGGUUCACCAUGGAAGCUCUUGAGGAGACCUGGCGUAACUUGCAAAAGAUCA .((((...((....)).(((..((.((((....(((((((((((..(((((((.....)))).)))(((((....))))))))))))))))(....))))))).....))).)))).... ( -40.10) >DroWil_CAF1 2671 120 - 1 ACUUUAAGGCAUUGGCAGCACUCGACCAGAAGAUCAAGAGCUUUAAUGUUGGACCGAAUCCAUACACAUGGUUCACCAUGGAAGCUCUUGAGGAGACCUGGCGUAAUCUGCAAAAGAUCA ..............((((....((.((((....(((((((((((..(((((((.....)))).)))(((((....))))))))))))))))(....)))))))....))))......... ( -41.80) >DroMoj_CAF1 2240 120 - 1 ACUUUAAGGCAUUGGCAGCACUCGACCAGAAGAUCAAGAGCUUUAAUGUUGGACCGAAUCCGUACACCUGGUUCACCAUGGAAGCUCUUGAGGAGACCUGGCGUAAUCUGCAAAAGAUCA ..............((((....((.((((....(((((((((((.(((.(((((((............))))))).))).)))))))))))(....)))))))....))))......... ( -41.10) >DroAna_CAF1 2024 120 - 1 AUUUCAAGGCAUUGGCAGCACUCGACCAGAAGAUCAAGAGCUUUAAUGUUGGACCCAAUCCCUACACCUGGUUCACCAUGGAAGCUCUUGAGGAGACCUGGCGUAACCUGCAAAAGAUCA ..............((((....((.((((....(((((((((((.(((.((((((..............)))))).))).)))))))))))(....)))))))....))))......... ( -39.84) >consensus ACUUUAAGGCAUUGGCAGCACUCGACCAGAAGAUCAAGAGCUUUAAUGUUGGACCCAAUCCGUACACCUGGUUCACCAUGGAAGCUCUUGAGGAGACCUGGCGUAAUCUGCAAAAGAUCA ..............((((....((.((((....(((((((((((.(((.((((((..............)))))).))).)))))))))))(....)))))))....))))......... (-39.88 = -39.74 + -0.14)

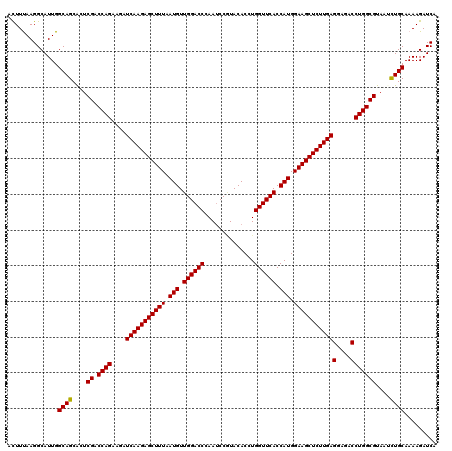
| Location | 1,764,864 – 1,764,984 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -40.77 |
| Consensus MFE | -38.26 |
| Energy contribution | -37.17 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1764864 120 - 23771897 GGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUCCGCGCCCUGCGCGACGCCCAUGCCCAAUUCCAGGCGUCCCUUUCGUCGGCCGAAGCUG (.((((....(((((((.((((((.....)))))).))))..)))..(((((.((((......((((...))))((((((..............)))))).)))).))))).)))).).. ( -42.94) >DroVir_CAF1 2023 120 - 1 GGUUCGAAAAUGCCGAGGAGGAUCUCACGGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCCCUGCGUGACGCGCACGCACAAUUCCAAGCGUCUCUCUCAUCGGCCGAAGCUG (.((((....(((((((.((((((.....)))))).))))..)))..(((((.(((((...((((....((((((.....)))))).........)))).))).))))))).)))).).. ( -38.12) >DroGri_CAF1 2515 120 - 1 GGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCCUUGCGUGAUGCCCACGCUCAAUUCCAAGCGUCCCUCUCAUCGGCCGAAGCUG (.((((.((((((((((.((((((.....)))))).))))..)).))))..(((((((...((((.....(((((.....)))))..........))))...)))..)))).)))).).. ( -37.66) >DroMoj_CAF1 2360 120 - 1 GGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCGCUGCGUGAUGCCCACGCCCAAUUCCAAGCGUCUCUUUCAUCGGCCGAAGCGG (((........)))(((.((((((.....)))))).))).((((..((((..(((.((((.....((((((((((.....))))).........)))))...)))).)))..)))))))) ( -40.90) >DroAna_CAF1 2144 120 - 1 GGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAACUCGAUCGAAGAAAUUCGCGCCCUGCGCGACGCCCACGCCCAAUUCCAGGCGUCCCUGUCGUCGGCCGAAGCUG (((.((((..(((((((.((((((.....)))))).))))..))).)))))))..((...((((.(((....((((((...(((((........)))))...)))))).))))))).)). ( -46.60) >DroPer_CAF1 2447 120 - 1 GGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCGUUGCGCGACGCCCACGCACAAUUCCAAGCGUCUCUGUCCUCGGCCGAAGCUG ((((((((..(((((((.((((((.....)))))).))))..))).))))).(((.((.....((((...))))(((((................)))))....)).))))))....... ( -38.39) >consensus GGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCCCUGCGCGACGCCCACGCCCAAUUCCAAGCGUCCCUCUCAUCGGCCGAAGCUG ((((((((..(((((((.((((((.....)))))).))))..))).))))).(((.((.....((((...))))(((((................)))))....)).))))))....... (-38.26 = -37.17 + -1.08)

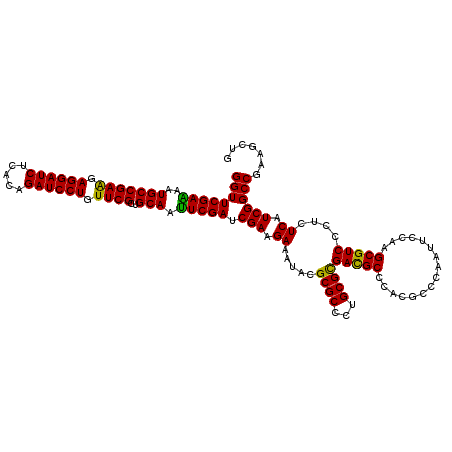
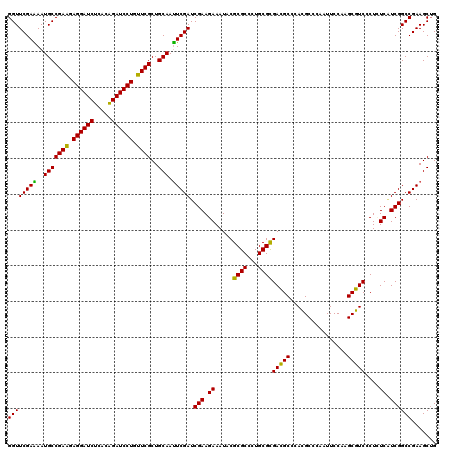
| Location | 1,764,904 – 1,765,024 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.61 |
| Mean single sequence MFE | -40.78 |
| Consensus MFE | -36.01 |
| Energy contribution | -36.62 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
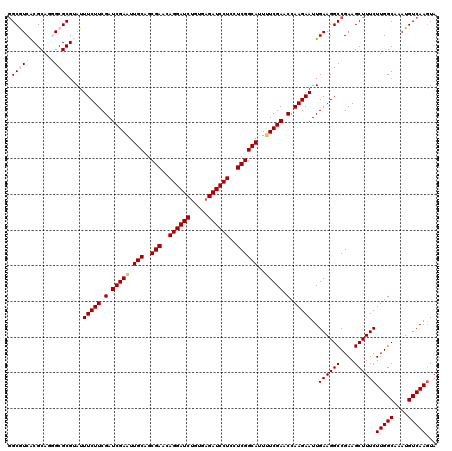
>3L_DroMel_CAF1 1764904 120 + 23771897 GGCGUCGCGCAGGGCGCGGAUUUCUUCGAUCGAAUUGCAGCGAACAGGAUCUGUGAGAUCCUCCUCGGCAUUUUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUA (((.((((((...))))))..(((((.(.(((((.(((..(((..((((((.....))))))..))))))..))))).).)))))......)))........((((((....)))))).. ( -41.50) >DroVir_CAF1 2063 120 + 1 CGCGUCACGCAGGGCGCGUAUUUCUUCGAUCGAAUUGCAGCGAACAGGAUCCGUGAGAUCCUCCUCGGCAUUUUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUA ((((((......))))))...(((((.(.(((((.(((..(((..(((((.(....))))))..))))))..))))).).)))))..((((((....))))))(((((....)))))... ( -40.20) >DroPse_CAF1 2465 120 + 1 GGCGUCGCGCAACGCGCGUAUUUCUUCGAUCGAAUUGCAGCGAACAGGAUCUGUGAGAUCCUCUUCGGCAUUUUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUA (((..(((((...)))))...(((((.(.(((((.(((..((((.((((((.....)))))).)))))))..))))).).)))))......)))........((((((....)))))).. ( -42.00) >DroGri_CAF1 2555 120 + 1 GGCAUCACGCAAGGCGCGUAUUUCUUCGAUCGAAUUGCAGCGAACAGGAUCUGUGAGAUCCUCUUCGGCAUUCUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUA (((.((((((.....))))..(((((.(.((((..(((..((((.((((((.....)))))).)))))))...)))).).)))))..))..)))........((((((....)))))).. ( -38.60) >DroMoj_CAF1 2400 120 + 1 GGCAUCACGCAGCGCGCGUAUUUCUUCGAUCGAAUUGCAGCGAACAGGAUCUGUGAGAUCCUCCUCGGCAUUUUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAAUA (((((.((((.....))))..(((((.(.(((((.(((..(((..((((((.....))))))..))))))..))))).).)))))..((((((....)))))).......)))))..... ( -36.30) >DroAna_CAF1 2184 120 + 1 GGCGUCGCGCAGGGCGCGAAUUUCUUCGAUCGAGUUGCAGCGAACAGGAUCUGUGAGAUCCUCUUCGGCAUUCUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUA (((.((((((...))))))..(((((.(.(((((.(((..((((.((((((.....)))))).)))))))..))))).).)))))......)))........((((((....)))))).. ( -46.10) >consensus GGCGUCACGCAGGGCGCGUAUUUCUUCGAUCGAAUUGCAGCGAACAGGAUCUGUGAGAUCCUCCUCGGCAUUUUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUA .(((((......)))))....(((((.(.(((((.(((..(((..((((((.....))))))..))))))..))))).).)))))..((((((....))))))(((((....)))))... (-36.01 = -36.62 + 0.61)

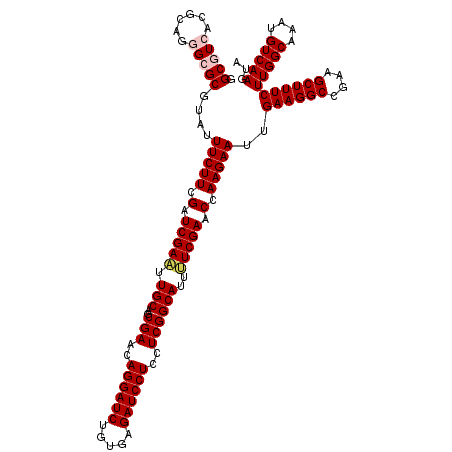
| Location | 1,764,904 – 1,765,024 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.61 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -41.79 |
| Energy contribution | -40.93 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.56 |
| Structure conservation index | 1.01 |
| SVM decision value | 5.59 |
| SVM RNA-class probability | 0.999990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
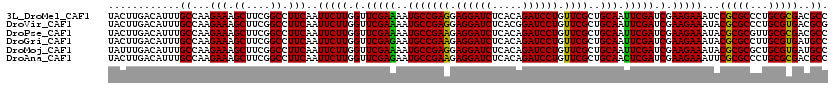
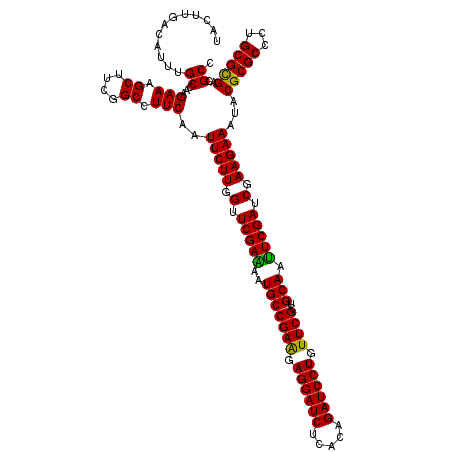
>3L_DroMel_CAF1 1764904 120 - 23771897 UACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUCCGCGCCCUGCGCGACGCC ............((...(((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...)))))..)). ( -41.30) >DroVir_CAF1 2063 120 - 1 UACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACGGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCCCUGCGUGACGCG ............((...(((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...)))))..)). ( -40.60) >DroPse_CAF1 2465 120 - 1 UACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCGUUGCGCGACGCC ............((...(((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...)))))..)). ( -41.80) >DroGri_CAF1 2555 120 - 1 UACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCCUUGCGUGAUGCC .................(((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...)))))..... ( -39.30) >DroMoj_CAF1 2400 120 - 1 UAUUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCGCUGCGUGAUGCC ..(((((.....((((((((.((....))......))))))))(((((..(((((((.((((((.....)))))).))))..))).))))))))))......(((((...)))))..... ( -39.50) >DroAna_CAF1 2184 120 - 1 UACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAACUCGAUCGAAGAAAUUCGCGCCCUGCGCGACGCC ............((...(((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))..((((((...)))))).)). ( -44.70) >consensus UACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCGCUGCAAUUCGAUCGAAGAAAUACGCGCCCUGCGCGACGCC ............((...(((.((....)).)))..(((((.(.(((((..(((((((.((((((.....)))))).))))..))).))))).).)))))...(((((...)))))..)). (-41.79 = -40.93 + -0.86)
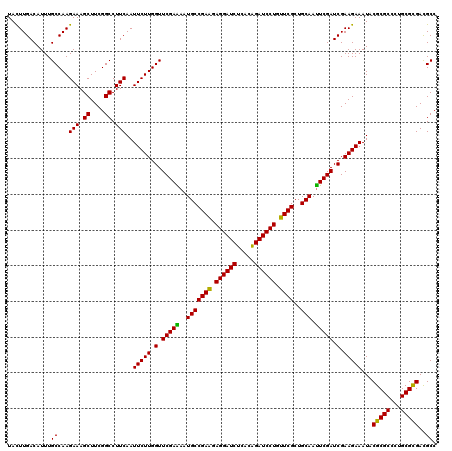
| Location | 1,764,944 – 1,765,064 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -36.15 |
| Energy contribution | -36.07 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1764944 120 + 23771897 CGAACAGGAUCUGUGAGAUCCUCCUCGGCAUUUUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUAGAGUUCCUCGAUCUGGCGGAACUGCUCCUGCAUGGCCAGC ....((((((((((.(((((....(((((...(((((........))))).)))))((((((((((((....)))))).))))))....))))).))))).....))))).......... ( -40.30) >DroVir_CAF1 2103 120 + 1 CGAACAGGAUCCGUGAGAUCCUCCUCGGCAUUUUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUAGAGUUCCUCGAUUUGGCGGAACUGCUCUUGCAUGGCCAGC .((.(((..(((((.(((((....(((((...(((((........))))).)))))((((((((((((....)))))).))))))....))))).))))).))).))............. ( -39.30) >DroGri_CAF1 2595 120 + 1 CGAACAGGAUCUGUGAGAUCCUCUUCGGCAUUCUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUAGAGUUCCUCGAUUUGGCGGAAUUGCUCCUGCAUGGCCAGC .(((.((((((.....)))))).)))(((((((.((..((((..(((((.((....((((((((((((....)))))).)))))))))))))))))))))))(((....)))..)))... ( -39.60) >DroMoj_CAF1 2440 120 + 1 CGAACAGGAUCUGUGAGAUCCUCCUCGGCAUUUUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAAUAGAGUUCCUCGAUUUGGCGGAAUUGCUCCUGCAUGGCCAGC (((..((((((.....))))))..)))((...(((((........)))))(((((..((......(((((.((((((((.(((...))).))))))))...)))))...)).))))).)) ( -36.00) >DroAna_CAF1 2224 120 + 1 CGAACAGGAUCUGUGAGAUCCUCUUCGGCAUUCUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUAGAGUUCCUCGAUCUGGCGGAACUGCUCCUGCAUUGCCAGA .((((((((((.....))))))(((((((.(((..............))).)))))))((..((((((....)))))).)).)))).....((((((((...(((....))))))))))) ( -42.54) >DroPer_CAF1 2527 120 + 1 CGAACAGGAUCUGUGAGAUCCUCUUCGGCAUUUUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUAGAGUUCCUCGAUUUGGCGGAACUGCUCNNNNNNNNNNNNN ((((.((((((.....)))))).))))(((.(((((..((((..(((((.((....((((((((((((....)))))).)))))))))))))))))))))).)))............... ( -38.20) >consensus CGAACAGGAUCUGUGAGAUCCUCCUCGGCAUUUUCGAACCAAGAAUUGAAGGCCGAAGCUUUCUUGGCAAAUGUCAAGUAGAGUUCCUCGAUUUGGCGGAACUGCUCCUGCAUGGCCAGC .((.(((..(((((.(((((....(((((...(((((........))))).)))))((((((((((((....)))))).))))))....))))).))))).))).))............. (-36.15 = -36.07 + -0.08)

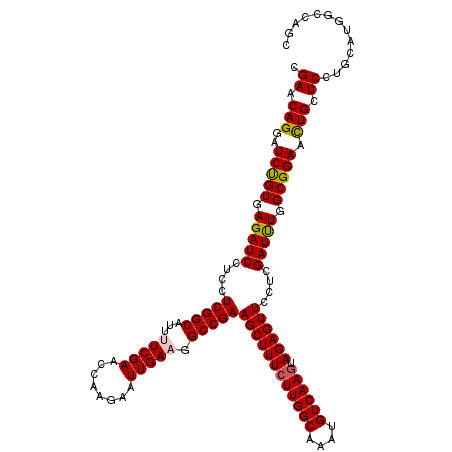
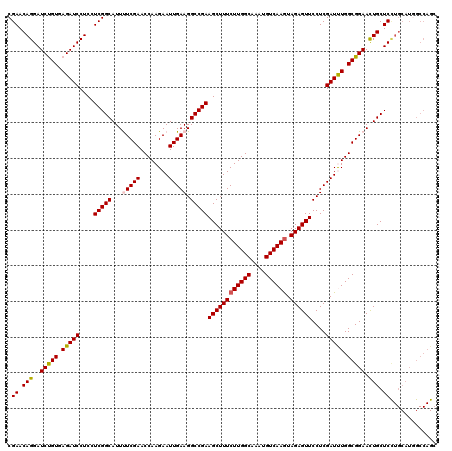
| Location | 1,764,944 – 1,765,064 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -35.48 |
| Energy contribution | -36.82 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1764944 120 - 23771897 GCUGGCCAUGCAGGAGCAGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCG ..((((.(((((((((.((((((..........)))))))).)))).)))..)))).(((.((....)).)))..((((((((........))))))))(((((.....)))))...... ( -42.30) >DroVir_CAF1 2103 120 - 1 GCUGGCCAUGCAAGAGCAGUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACGGAUCCUGUUCG ..((((.(((((((((.((((((..........)))))))).)))).)))..)))).(((.((....)).)))..((((((((........))))))))(((((.....)))))...... ( -42.70) >DroGri_CAF1 2595 120 - 1 GCUGGCCAUGCAGGAGCAAUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCG ((((((.(((((((((...((((..........))))..)).)))).)))..)))).(((.((....)).))).(((((((...)))))))))((((.((((((.....)))))).)))) ( -38.70) >DroMoj_CAF1 2440 120 - 1 GCUGGCCAUGCAGGAGCAAUUCCGCCAAAUCGAGGAACUCUAUUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAGGAGGAUCUCACAGAUCCUGUUCG ...(((......((((...((((..........)))))))).(((((.....((((((((.((....))......)))))))))))))...)))(((.((((((.....)))))).))). ( -36.10) >DroAna_CAF1 2224 120 - 1 UCUGGCAAUGCAGGAGCAGUUCCGCCAGAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAGAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCG ..((((((((((((((.((((((..........)))))))).)))).)).)))))).(((.((....)).))).(((((((...)))))))..((((.((((((.....)))))).)))) ( -46.40) >DroPer_CAF1 2527 120 - 1 NNNNNNNNNNNNNGAGCAGUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCG ..............((.((((((..........))))))))......(((((((((((((.((....))......))))))))....))))).((((.((((((.....)))))).)))) ( -34.40) >consensus GCUGGCCAUGCAGGAGCAGUUCCGCCAAAUCGAGGAACUCUACUUGACAUUUGCCAAGAAAGCUUCGGCCUUCAAUUCUUGGUUCGAAAAUGCCGAAGAGGAUCUCACAGAUCCUGUUCG ...(((.(((((((((.((((((..........)))))))).)))).)))..((((((((.((....))......))))))))........)))(((.((((((.....)))))).))). (-35.48 = -36.82 + 1.34)
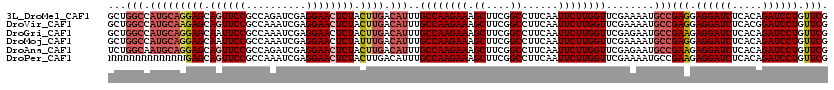
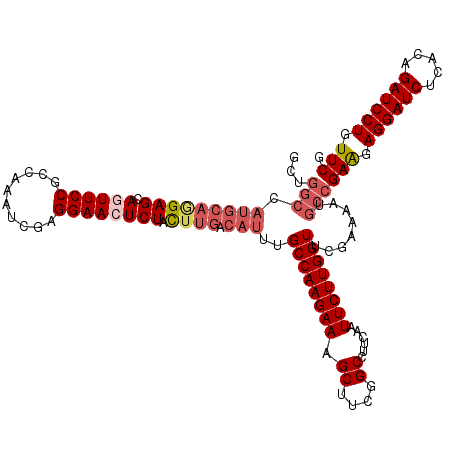
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:35 2006