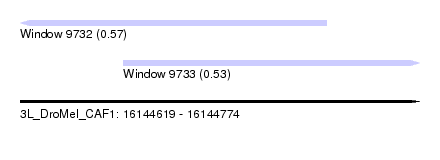
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,144,619 – 16,144,774 |
| Length | 155 |
| Max. P | 0.568224 |

| Location | 16,144,619 – 16,144,738 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -28.28 |
| Energy contribution | -28.46 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16144619 119 - 23771897 UGAACCCCUUCUCCUGCCGGCAAGCAAAAAUAGGCGGCUGACGUCAAAAUGCAAGCCAGACGGGAAAAA-GGGGAGGGGUGAGUGGGGAAAGAAACCGAAAAAAAACCGAGUUGGAGUCG ....((((((.((((((.(((..(((......((((.....))))....)))..))).).)))))..))-)))).((..(...(.((........)).)....)..))............ ( -34.50) >DroSec_CAF1 6657 115 - 1 UGAGCCCCUUCUGCUGC-----AACAAAAAUAGGCGGCUGACGUCAAAAUGCAAGCCAGACGGGAAAUGGGGGGAGGGGUGAGUGGGGAACGAAACCGAAAAAAAACCAAGUUGGAGUCG ...(((((((((.((((-----...........))((((..(((....)))..))))...........)).)))))))))..........(((..((((............))))..))) ( -30.10) >DroSim_CAF1 7455 113 - 1 UGAACCCCUUCUCCUGC-----AACAAAAAUAGGCGGCUGACGUCAAAAUGCAAGCCAGACGGGAAAUGGGGGGAGGGGUGAGUGGGGAACGAAACCGAAA--AAACCAAGUUGGAGUCG ...((((((((((((((-----...........))((((..(((....)))..))))...........))))))))))))..........(((..((((..--........))))..))) ( -33.80) >DroEre_CAF1 6215 111 - 1 UUGACCCCUCCCCCUGC-----AACAAAAAUAGGCGGCUGACGUCAAAAUGCAAGCCAGACGGGUAAAA-AGGGAGGGGUGUGUGGGGAACGAAACCGAG---AAACCAAGUUGGAGUCG ...(((((((((((((.-----........)))).((((..(((....)))..))))............-.))))))))).....((........))..(---(..((.....))..)). ( -33.10) >consensus UGAACCCCUUCUCCUGC_____AACAAAAAUAGGCGGCUGACGUCAAAAUGCAAGCCAGACGGGAAAAA_GGGGAGGGGUGAGUGGGGAACGAAACCGAAA__AAACCAAGUUGGAGUCG ...((((((((((((((................))((((..(((....)))..))))...........))))))))))))...........((..((((............))))..)). (-28.28 = -28.46 + 0.19)


| Location | 16,144,659 – 16,144,774 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.64 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -24.76 |
| Energy contribution | -24.57 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16144659 115 + 23771897 ACCCCUCCCC-UUUUUCCCGUCUGGCUUGCAUUUUGACGUCAGCCGCCUAUUUUUGCUUGCCGGCAGGAGAAGGGGUUCAAAAGAAUCGAGCAGACAUAAAAAAAC----ACGAGAAAAA ....(((.((-(((((((.(.(((((..(((....(.((.....)).)......)))..)))))).)))))))))((((.........))))..............----..)))..... ( -30.70) >DroSec_CAF1 6697 115 + 1 ACCCCUCCCCCCAUUUCCCGUCUGGCUUGCAUUUUGACGUCAGCCGCCUAUUUUUGUU-----GCAGCAGAAGGGGCUCAAAAGAAUCGGCCAGACAUUUAAAAAAAGUAAAAAAAAAAA ...................((((((((....((((((.....(((.(((...(((((.-----...))))))))))))))))).....))))))))........................ ( -28.00) >DroSim_CAF1 7493 103 + 1 ACCCCUCCCCCCAUUUCCCGUCUGGCUUGCAUUUUGACGUCAGCCGCCUAUUUUUGUU-----GCAGGAGAAGGGGUUCAAAAGAAUCGACCAGACAU------------AAAAAAAAUA ...................((((((.(((..((((((.....(((.(((.((((((..-----.)))))).))))))))))))....)))))))))..------------.......... ( -23.20) >DroEre_CAF1 6252 107 + 1 ACCCCUCCCU-UUUUACCCGUCUGGCUUGCAUUUUGACGUCAGCCGCCUAUUUUUGUU-----GCAGGGGGAGGGGUCAAAAGGAAUCGACCAGACAUAAAAAGGC----AACCAAA--- .......(((-(((((...((((((.(((..(((((((.((..((.(((.........-----..))).))..))))))))).....))))))))).)))))))).----.......--- ( -31.50) >consensus ACCCCUCCCC_CAUUUCCCGUCUGGCUUGCAUUUUGACGUCAGCCGCCUAUUUUUGUU_____GCAGGAGAAGGGGUUCAAAAGAAUCGACCAGACAUAAAAAAAC____AAAAAAAAAA ...................((((((.(((.(((((......((((.(((.(((((((......))))))).)))))))....))))))))))))))........................ (-24.76 = -24.57 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:57 2006