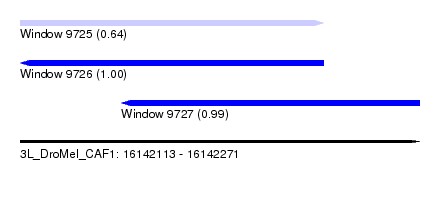
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,142,113 – 16,142,271 |
| Length | 158 |
| Max. P | 0.995826 |

| Location | 16,142,113 – 16,142,233 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -20.99 |
| Consensus MFE | -18.16 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16142113 120 + 23771897 ACGAGAAAAUAACCGAAGAAAAAGUCGAACAAAGCUUAAGAAAAGAACACUUGUAUGUACCUUCUCUGCAAAGGAUACACAAGUGUCUCAUUAAAAGAAAUAAAGGACAAGCCAAGCGCC .............(((........)))......((((.......((((((((((.(((.((((.......))))))).)))))))).))...............((.....))))))... ( -21.10) >DroSec_CAF1 4319 120 + 1 ACGAUAAAAUAACCGGAGAAUAAGUCGAACAAAGCUUAAGAAAAGAACACUUGUAUGUACCUUCUCUGCAAAGGAUACACAAGUGUCUCAUUAAAAGAAAUAAAGGACAAGCCAAGCGCC .((((..................))))......((((.......((((((((((.(((.((((.......))))))).)))))))).))...............((.....))))))... ( -21.37) >DroSim_CAF1 5089 120 + 1 ACGAGAAAAUAACCGGAGAAAAAGUCGAACAAAGCUUAAGAAAAGAACACUUGUAUGUACGUUCUCUGCAAAGGAUACACAAGUGUCUCAUUAAAAGAAAUAAAGGACAAGCCAAGCGCC ..............((((((...((((.((((..(((.....))).....)))).)).)).))))))((...((((((....))))))................((.....))..))... ( -17.70) >DroEre_CAF1 3616 120 + 1 ACGAGAAAAUAACCUGAGAAAAAGUCGAACAAAGCUUAAGAAAAGGACACUUGUAUGUACCUUCUCUGCAAAGGAUACACAAGUGUCUCAUUAAAAGAAAUAAAGGACAAGCCAAGCGCC .................................((((.......((((((((((.(((.((((.......))))))).))))))))))................((.....))))))... ( -23.20) >DroYak_CAF1 4332 120 + 1 ACGAGAAAAUAACCUGAGAAAAAGUCGAACAAAGCAUAAGAAAAGGACACUUGUAUGUACCUUCUCUGCAAAGGAUACACAAGUGUCUCAUUAAAAGAAAUAAAGGACAAGCCAAGCGCC .................................((.........((((((((((.(((.((((.......))))))).))))))))))................((.....))..))... ( -21.60) >consensus ACGAGAAAAUAACCGGAGAAAAAGUCGAACAAAGCUUAAGAAAAGAACACUUGUAUGUACCUUCUCUGCAAAGGAUACACAAGUGUCUCAUUAAAAGAAAUAAAGGACAAGCCAAGCGCC .................................((((.......((((((((((.(((.((((.......))))))).)))))))).))...............((.....))))))... (-18.16 = -18.32 + 0.16)



| Location | 16,142,113 – 16,142,233 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16142113 120 - 23771897 GGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAAGGUACAUACAAGUGUUCUUUUCUUAAGCUUUGUUCGACUUUUUCUUCGGUUAUUUUCUCGU .(.((..((((((....(((((......)))))(((((((((((..(((((.....)))))...))))))))))).......))))))..)).)((((........)))).......... ( -26.70) >DroSec_CAF1 4319 120 - 1 GGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAAGGUACAUACAAGUGUUCUUUUCUUAAGCUUUGUUCGACUUAUUCUCCGGUUAUUUUAUCGU .(.((..((((((....(((((......)))))(((((((((((..(((((.....)))))...))))))))))).......))))))..)).)...........((((......)))). ( -27.70) >DroSim_CAF1 5089 120 - 1 GGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAACGUACAUACAAGUGUUCUUUUCUUAAGCUUUGUUCGACUUUUUCUCCGGUUAUUUUCUCGU .(.((..((((((....(((((......)))))(((((((((((((.(((....)))...))))..))))))))).......))))))..)).)((((........)))).......... ( -20.40) >DroEre_CAF1 3616 120 - 1 GGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAAGGUACAUACAAGUGUCCUUUUCUUAAGCUUUGUUCGACUUUUUCUCAGGUUAUUUUCUCGU .(.((..((((((....(((((......)))))(((((((((((..(((((.....)))))...))))))))))).......))))))..)).)(((((......))))).......... ( -29.70) >DroYak_CAF1 4332 120 - 1 GGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAAGGUACAUACAAGUGUCCUUUUCUUAUGCUUUGUUCGACUUUUUCUCAGGUUAUUUUCUCGU .(.((..(((.......(((((......)))))(((((((((((..(((((.....)))))...)))))))))))..........)))..)).)(((((......))))).......... ( -26.60) >consensus GGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAAGGUACAUACAAGUGUUCUUUUCUUAAGCUUUGUUCGACUUUUUCUCCGGUUAUUUUCUCGU .(.((..((((((....(((((......)))))(((((((((((..(((((.....)))))...))))))))))).......))))))..)).)((((........)))).......... (-25.94 = -26.10 + 0.16)


| Location | 16,142,153 – 16,142,271 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.90 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -31.22 |
| Energy contribution | -31.18 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16142153 118 - 23771897 GUCAGCCAGGGAUUCGAGU--CGACUUUAAGGACUUCGUCGGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAAGGUACAUACAAGUGUUCUUUU ...((((((((...(((((--(.........))).)))....).)))))))....................(((((((((((((..(((((.....)))))...))))))))).)))).. ( -33.50) >DroSec_CAF1 4359 120 - 1 GUCUGCCAGGGAUUCGAGUCCCGACUUUAAGGACUUCGUCGGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAAGGUACAUACAAGUGUUCUUUU .......((((((((((((.(((((............))))).))))))...)))))).............(((((((((((((..(((((.....)))))...))))))))).)))).. ( -36.70) >DroSim_CAF1 5129 120 - 1 GUCUGCCAGGGAUUCGAGUCUCGACUUUAAGGACUUCGUCGGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAACGUACAUACAAGUGUUCUUUU .(((((.((((((.(((((.((.......(((((...((((.....))))..)))))(((((......))))))).)))))...)))))).)))))(((((..........))))).... ( -32.40) >DroEre_CAF1 3656 118 - 1 GAAUGCCAGGGAUUCGAGU--CGACUUUAAGGACUUCGUCGGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAAGGUACAUACAAGUGUCCUUUU ((..(((((((...(((((--(.........))).)))....).))))))...))..(((((......)))))(((((((((((..(((((.....)))))...)))))))))))..... ( -35.30) >DroYak_CAF1 4372 118 - 1 GACUGCCAGGGAUUCGAGU--CGACUUUAAGGACUUCGUCGGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAAGGUACAUACAAGUGUCCUUUU (((.(((((((...(((((--(.........))).)))....).))))))..)))..(((((......)))))(((((((((((..(((((.....)))))...)))))))))))..... ( -37.50) >consensus GUCUGCCAGGGAUUCGAGU__CGACUUUAAGGACUUCGUCGGCGCUUGGCUUGUCCUUUAUUUCUUUUAAUGAGACACUUGUGUAUCCUUUGCAGAGAAGGUACAUACAAGUGUUCUUUU .......((((((((((((..((((............))))..))))))...))))))...............(((((((((((..(((((.....)))))...)))))))))))..... (-31.22 = -31.18 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:50 2006