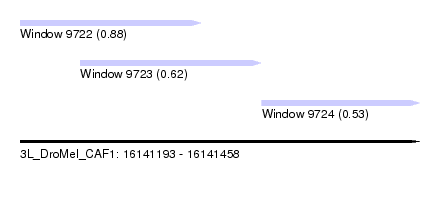
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,141,193 – 16,141,458 |
| Length | 265 |
| Max. P | 0.883910 |

| Location | 16,141,193 – 16,141,313 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -40.90 |
| Energy contribution | -41.03 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16141193 120 + 23771897 GCACAUCUGCAAGCAGCCUAUCAAUUUUUGACGAGACAAUCUCAGAGUCCAAAGGGGCACUGCAAAAAAAUGCAGUCGGAGCAAUUGGCUGGCGGAUCAGGGGUCGCUCGAGAGGUGAGC ((.(((((.(.(((.(((..(((((((((((((((.....)))...((((....))))..((((......))))))))))..))))))..))).(((.....)))))).)..))))).)) ( -37.90) >DroSec_CAF1 3460 120 + 1 GCACAUCUGCAAGCAGCCUAUCAAUUUUUGACGAGACAAUCUCAGAGUCCAAAGGGGCACUGCAAAAAAAUGCAGUCGGAGCAAUUGGCUGGCGGACCAGGGGUCGCUCGAGAGGUGAGC ((.(((((.(.(((.(((..(((((((((((((((.....)))...((((....))))..((((......))))))))))..))))))..))).((((...))))))).)..))))).)) ( -40.20) >DroEre_CAF1 2823 119 + 1 GCACAUCUGCAAGCAGCCUAUCAAUUUUUGACGAGACAAUCUCAGAGUCCAAAGGGGCACUGCAAA-AAAUGCAGUCGGAGCAAUUGGCUGGCGGACCAGGGGUCGCUUGAGAGGUGAGC ((.(((((.(((((.(((..(((((((((((((((.....)))...((((....))))..((((..-...))))))))))..))))))..))).((((...)))))))))..))))).)) ( -44.10) >DroYak_CAF1 3482 119 + 1 UCACAUCUGCAAGCAGCCUAUCAAUUUUUGACGAGACAAUCUCAGAGUCCAAAGGGGCACUGCAAA-AAUUGCAGUCGGGGCAAUUGGCUGGCGGACCAGGGGUCGCUUGAGAGGUGAGC ((((.(((.(((((.(((..(((((((((((((((.....)))...((((....))))..((((..-...))))))))))..))))))..))).((((...)))))))))))).)))).. ( -43.80) >consensus GCACAUCUGCAAGCAGCCUAUCAAUUUUUGACGAGACAAUCUCAGAGUCCAAAGGGGCACUGCAAA_AAAUGCAGUCGGAGCAAUUGGCUGGCGGACCAGGGGUCGCUCGAGAGGUGAGC .(((.(((.(((((.(((..(((((((((((((((.....)))...((((....))))..((((......))))))))))..))))))..))).((((...)))))))))))).)))... (-40.90 = -41.03 + 0.12)



| Location | 16,141,233 – 16,141,353 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.73 |
| Mean single sequence MFE | -44.62 |
| Consensus MFE | -39.15 |
| Energy contribution | -38.77 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16141233 120 + 23771897 CUCAGAGUCCAAAGGGGCACUGCAAAAAAAUGCAGUCGGAGCAAUUGGCUGGCGGAUCAGGGGUCGCUCGAGAGGUGAGCUCACUUCGGCCAGGGGCAUGGGGCAGCAGCAGCGCUGCAA ((((..((((......((((((((......))))))....))......(((((.((...(((.((((.(....))))).)))...)).))))))))).))))(((((......))))).. ( -49.40) >DroSec_CAF1 3500 114 + 1 CUCAGAGUCCAAAGGGGCACUGCAAAAAAAUGCAGUCGGAGCAAUUGGCUGGCGGACCAGGGGUCGCUCGAGAGGUGAGCUUACUGCGGCCAGGGGCGUGGGGCA------GUGCUGCAA ......((((....))))((((((......)))))).(.((((.(((.((.(((..((...((((((.....(((....)))...)))))).))..))).)).))------))))).).. ( -44.90) >DroEre_CAF1 2863 113 + 1 CUCAGAGUCCAAAGGGGCACUGCAAA-AAAUGCAGUCGGAGCAAUUGGCUGGCGGACCAGGGGUCGCUUGAGAGGUGAGCUCAGUGCGGCCAGGGGCGUGGGGCA------GCACUGCAA ..(((.((((....)))).)))....-...((((((....((...((.((.(((..((...(((((((((((.......))))).)))))).))..))).)).))------)))))))). ( -43.70) >DroYak_CAF1 3522 113 + 1 CUCAGAGUCCAAAGGGGCACUGCAAA-AAUUGCAGUCGGGGCAAUUGGCUGGCGGACCAGGGGUCGCUUGAGAGGUGAGCUCACUGUGGCCAGGGGCGUGGGGAA------UCGCUACAA (((...((((....))))((((((..-...)))))).)))((..(((((((.(((....(((.(((((.....))))).))).))))))))))..))((((.(..------.).)))).. ( -40.50) >consensus CUCAGAGUCCAAAGGGGCACUGCAAA_AAAUGCAGUCGGAGCAAUUGGCUGGCGGACCAGGGGUCGCUCGAGAGGUGAGCUCACUGCGGCCAGGGGCGUGGGGCA______GCGCUGCAA ......((((....))))((((((......))))))....((..(((((((.(((....(((.(((((.....))))).))).))))))))))..))((((.((.......)).)))).. (-39.15 = -38.77 + -0.38)


| Location | 16,141,353 – 16,141,458 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.85 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -28.75 |
| Energy contribution | -29.06 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16141353 105 + 23771897 GAGGGGCAGGGGGGUGUUUGAGGGGAUGGGGGCUGGGCAGACGUCUGCGGUCUGUGCAUAAAAUUACGAUAAGUGCAGGCGAGGCCACAGUCCAGAGGGCUCAGA ...........((((.(((....((((...((((..((((....)))).(((((..(...............)..)))))..))))...)))).))).))))... ( -34.76) >DroSec_CAF1 3614 102 + 1 GAGGGGCAGG-GGGUGUUUGAGGGGAUGGGGGCCGGGCAGACGUCUGCGGUCUGUGCAUGAAAUUACGAUAAGUGCAGGCGAGGCCGCAGUCCAGAGGGC--AGA ..........-...(((((....((((.(.((((..((((....)))).(((((..(...............)..)))))..)))).).))))...))))--).. ( -35.06) >DroEre_CAF1 2976 101 + 1 GGGGGGGCAGG-GGUGUUUGAAGGGAAG-GGGCUGGGCAGACGUCUGCGGUCUGUGCAUAAAAUUACGAUAAGUGCAGGCGAGGCCGCAGUCCAGAGGGC--AGA ....(((((..-..))))).........-(((((((((...(((((((((((.(((........))))))...))))))))..))).)))))).......--... ( -30.40) >DroYak_CAF1 3635 102 + 1 UCGGGGACAGGAGGUGUUUGAAGGGAAG-GGGCUGGGCAGACGUCUGCGGUCUGUGCAUAAAAUUACGAUAAGUGCAGGCGAGGCCACAGUCCAGAGGGC--AGA (((((.((.....)).))))).......-(((((((((...(((((((((((.(((........))))))...))))))))..))).)))))).......--... ( -33.10) >consensus GAGGGGCAAGGGGGUGUUUGAAGGGAAG_GGGCUGGGCAGACGUCUGCGGUCUGUGCAUAAAAUUACGAUAAGUGCAGGCGAGGCCACAGUCCAGAGGGC__AGA .............((.(((....((((...((((..((((....)))).(((((..(...............)..)))))..))))...)))).))).))..... (-28.75 = -29.06 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:47 2006