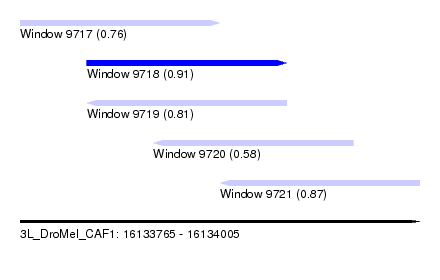
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,133,765 – 16,134,005 |
| Length | 240 |
| Max. P | 0.910157 |

| Location | 16,133,765 – 16,133,885 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -32.07 |
| Energy contribution | -32.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16133765 120 + 23771897 CCAAUACAAAGAGCCCUGUCUCGAAUCAUAACUCAUUGAACUUUUGCCGGCAGCAGCAGCCGCUGCUGAAACAUAAAAAGUGUCAAUCAGUGACUCUGCUGUGGCAACGAUGGCGACGCC ............((..(((((((..(((........)))....((((((.((((((.((.(((((.(((.((.......)).)))..))))).))))))))))))))))).))))..)). ( -36.20) >DroSim_CAF1 9071 117 + 1 CCAAUACAAAGAGCCCUGUCUCGAAUCAUAACUCAUUGAACUUUUGCCGGCAGCAGCAGCCGC---UGAAACAUAAAAAGUGUCAAUCAGUGACUCUGCUGUGGCAACGAUGGCGACGCC ............((..(((((((..(((........)))....((((((.((((((.((.(((---(((.((((.....))))...)))))).))))))))))))))))).))))..)). ( -37.50) >DroYak_CAF1 1958 117 + 1 CCAAUACAAAGAGCCCUGUCUCGAAUCAUAACUCAUUGAACUUUUGCCGGCAGCAGCAGCCGC---UGAAACAUAAAAAGUGUCAAUCAGUGACUCUGCUGUGGCAACGAUGGCGACGCC ............((..(((((((..(((........)))....((((((.((((((.((.(((---(((.((((.....))))...)))))).))))))))))))))))).))))..)). ( -37.50) >consensus CCAAUACAAAGAGCCCUGUCUCGAAUCAUAACUCAUUGAACUUUUGCCGGCAGCAGCAGCCGC___UGAAACAUAAAAAGUGUCAAUCAGUGACUCUGCUGUGGCAACGAUGGCGACGCC ............((..(((((((..(((........)))....((((((.((((((.((.(((...(((.((.......)).)))....))).))))))))))))))))).))))..)). (-32.07 = -32.07 + 0.00)

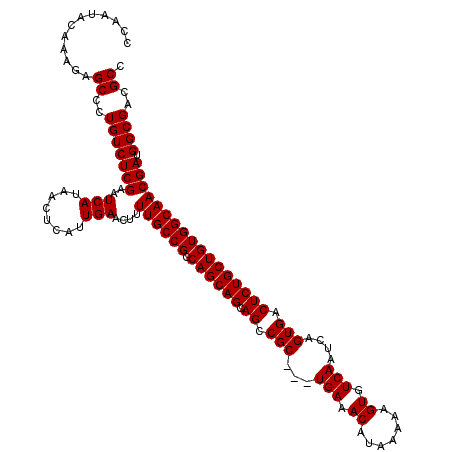
| Location | 16,133,805 – 16,133,925 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -35.57 |
| Energy contribution | -35.57 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16133805 120 + 23771897 CUUUUGCCGGCAGCAGCAGCCGCUGCUGAAACAUAAAAAGUGUCAAUCAGUGACUCUGCUGUGGCAACGAUGGCGACGCCUCCAAAGUCCCAUUACAAUGCCAUUUUUAUGGACACCGCU .....((.(((((((((....)))))))...(((((((((.((((.....)))).))...((((((..(((((.(((.........))))))))....)))))))))))))....)))). ( -39.80) >DroSim_CAF1 9111 117 + 1 CUUUUGCCGGCAGCAGCAGCCGC---UGAAACAUAAAAAGUGUCAAUCAGUGACUCUGCUGUGGCAACGAUGGCGACGCCUCCAAAGUCCCAUUACAAUGCCAUUUUUAUGGACACCGCU ...((((((.((((((.((.(((---(((.((((.....))))...)))))).)))))))))))))).(((((.(((.........))))))))....((((((....)))).))..... ( -41.00) >DroYak_CAF1 1998 117 + 1 CUUUUGCCGGCAGCAGCAGCCGC---UGAAACAUAAAAAGUGUCAAUCAGUGACUCUGCUGUGGCAACGAUGGCGACGCCUCCAAAGUCCCAUUACAAUGCCAUUUUUAUGGACACCGCU ...((((((.((((((.((.(((---(((.((((.....))))...)))))).)))))))))))))).(((((.(((.........))))))))....((((((....)))).))..... ( -41.00) >consensus CUUUUGCCGGCAGCAGCAGCCGC___UGAAACAUAAAAAGUGUCAAUCAGUGACUCUGCUGUGGCAACGAUGGCGACGCCUCCAAAGUCCCAUUACAAUGCCAUUUUUAUGGACACCGCU ...((((((.((((((.((.(((...(((.((.......)).)))....))).)))))))))))))).(((((.(((.........))))))))....((((((....)))).))..... (-35.57 = -35.57 + 0.00)

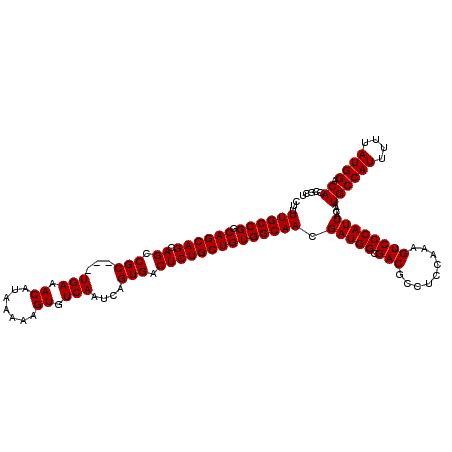
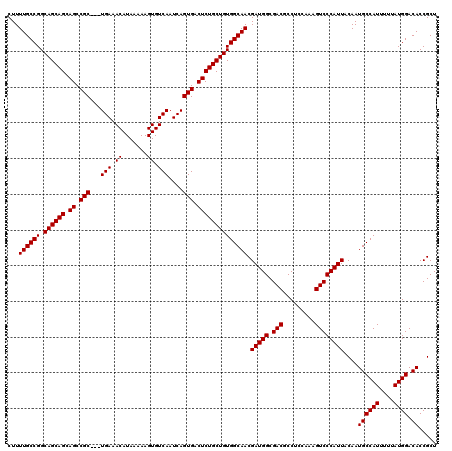
| Location | 16,133,805 – 16,133,925 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -43.57 |
| Consensus MFE | -38.47 |
| Energy contribution | -38.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16133805 120 - 23771897 AGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGAGGCGUCGCCAUCGUUGCCACAGCAGAGUCACUGAUUGACACUUUUUAUGUUUCAGCAGCGGCUGCUGCUGCCGGCAAAAG .((((((.((((....)))))))))).(((((((.........))).))))..(((((.((((((((((.(((.((((.((.......)).))))))).)))).))))))..)))))... ( -44.90) >DroSim_CAF1 9111 117 - 1 AGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGAGGCGUCGCCAUCGUUGCCACAGCAGAGUCACUGAUUGACACUUUUUAUGUUUCA---GCGGCUGCUGCUGCCGGCAAAAG .((((((.((((....)))))))))).(((((((.........))).))))..(((((.((((((((((.((((..((((.......)))))))---).)))).))))))..)))))... ( -42.90) >DroYak_CAF1 1998 117 - 1 AGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGAGGCGUCGCCAUCGUUGCCACAGCAGAGUCACUGAUUGACACUUUUUAUGUUUCA---GCGGCUGCUGCUGCCGGCAAAAG .((((((.((((....)))))))))).(((((((.........))).))))..(((((.((((((((((.((((..((((.......)))))))---).)))).))))))..)))))... ( -42.90) >consensus AGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGAGGCGUCGCCAUCGUUGCCACAGCAGAGUCACUGAUUGACACUUUUUAUGUUUCA___GCGGCUGCUGCUGCCGGCAAAAG .((((((.((((....)))))))))).(((((((.........))).))))..(((((.((((((((((.(....(((.((.......)).)))...).)))).))))))..)))))... (-38.47 = -38.47 + -0.00)


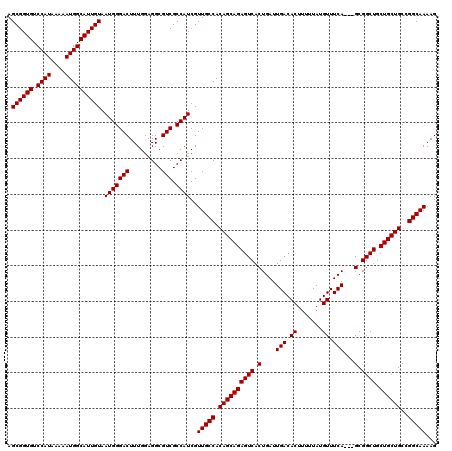
| Location | 16,133,845 – 16,133,965 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -42.83 |
| Consensus MFE | -42.83 |
| Energy contribution | -42.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16133845 120 - 23771897 GAGCUGCUCACUCCGCUCCAGCUGGAGCAGCAUCCCCGCCAGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGAGGCGUCGCCAUCGUUGCCACAGCAGAGUCACUGAUUGACA ...(((((...((((((((....)))))......((((...((((((.((((....))))))))))..)))).....)))((((.((....)).))))..))))).((((.....)))). ( -42.90) >DroSim_CAF1 9148 120 - 1 GAGCUGCUCACUCCGCUCCAGCUGGAGCAGCAUCCCCGCCAGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGAGGCGUCGCCAUCGUUGCCACAGCAGAGUCACUGAUUGACA ...(((((...((((((((....)))))......((((...((((((.((((....))))))))))..)))).....)))((((.((....)).))))..))))).((((.....)))). ( -42.90) >DroYak_CAF1 2035 117 - 1 ---CUGCUCACUCCGCUCCAGCUGGAGCAGCAUCCCCGCCAGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGAGGCGUCGCCAUCGUUGCCACAGCAGAGUCACUGAUUGACA ---(((((...((((((((....)))))......((((...((((((.((((....))))))))))..)))).....)))((((.((....)).))))..))))).((((.....)))). ( -42.70) >consensus GAGCUGCUCACUCCGCUCCAGCUGGAGCAGCAUCCCCGCCAGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGAGGCGUCGCCAUCGUUGCCACAGCAGAGUCACUGAUUGACA ...(((((...((((((((....)))))......((((...((((((.((((....))))))))))..)))).....)))((((.((....)).))))..))))).((((.....)))). (-42.83 = -42.83 + 0.00)

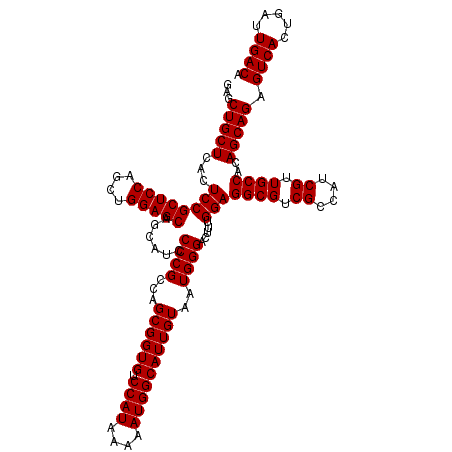
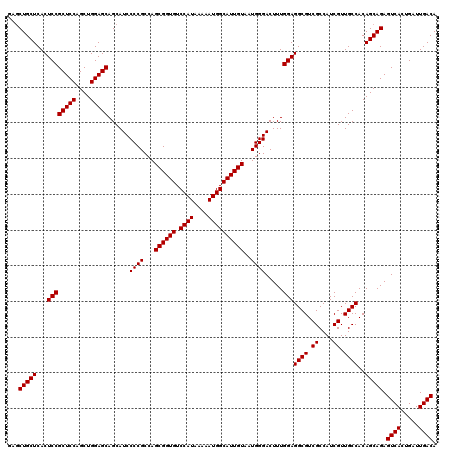
| Location | 16,133,885 – 16,134,005 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -28.67 |
| Energy contribution | -29.00 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16133885 120 - 23771897 CUCCUGCUCCUUCUACUACUCCACUCCUCGUCUCCAGAUGGAGCUGCUCACUCCGCUCCAGCUGGAGCAGCAUCCCCGCCAGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGA ...................((((......(.((((((.((((((..........)))))).)))))))......((((...((((((.((((....))))))))))..))))....)))) ( -38.30) >DroSim_CAF1 9188 120 - 1 CUCCUGCUCCUUCUACUCCUGCACUCCUCGUCUCCAGAUGGAGCUGCUCACUCCGCUCCAGCUGGAGCAGCAUCCCCGCCAGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGA .(((((((............(((((((((.......)).)))).))).......(((((....)))))))))..((((...((((((.((((....))))))))))..)))).....))) ( -38.90) >DroYak_CAF1 2075 92 - 1 GUCCUUCUCCUUCUU----------------------------CUGCUCACUCCGCUCCAGCUGGAGCAGCAUCCCCGCCAGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGA .(((...........----------------------------.((((..((((((....)).)))).))))..((((...((((((.((((....))))))))))..)))).....))) ( -27.30) >consensus CUCCUGCUCCUUCUACU_CU_CACUCCUCGUCUCCAGAUGGAGCUGCUCACUCCGCUCCAGCUGGAGCAGCAUCCCCGCCAGCGGUGUCCAUAAAAAUGGCAUUGUAAUGGGACUUUGGA .(((......................................(((((((.(...((....)).))))))))...((((...((((((.((((....))))))))))..)))).....))) (-28.67 = -29.00 + 0.33)

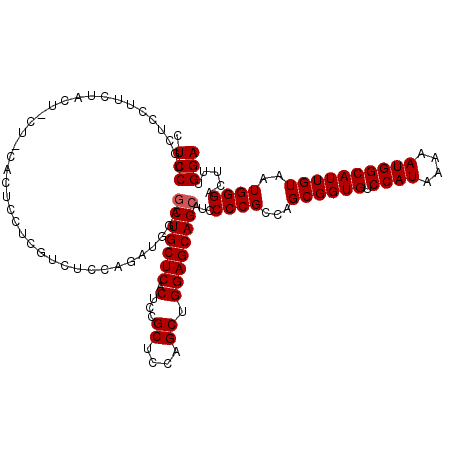
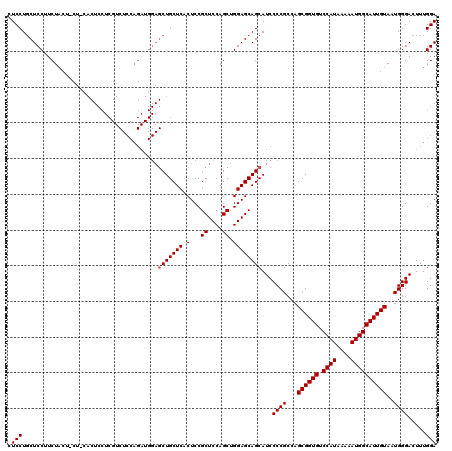
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:44 2006