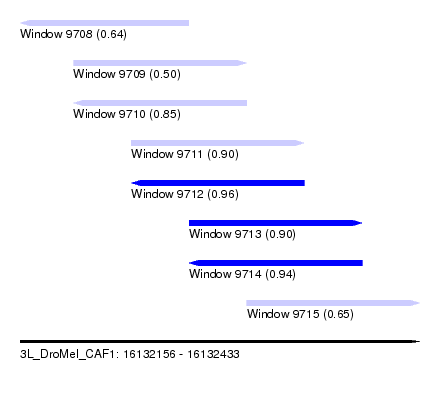
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,132,156 – 16,132,433 |
| Length | 277 |
| Max. P | 0.956822 |

| Location | 16,132,156 – 16,132,273 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.36 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.23 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16132156 117 - 23771897 UCCGAUGAAUCGAACGGCCAGGUGGCACAAAGCAGCCGCCAAAAAGUCAACAGAAAGCAAAUUGCCACCGCGAAUAGCCAACCUGAGACCCACAAAUCGUCUUUCCCCCA--CAU-CAAA ...((((....(((.(((..((((((........)))))).....(((..(((...((...((((....))))...))....))).))).........))).))).....--)))-)... ( -25.80) >DroSim_CAF1 7383 117 - 1 UCCGAUGAAUCAAACGGCCAGGUGGCUCAAAGCAGCCGCCAAAAAGUCAACAGAAAGCAAUUUGCCACCGCGAAUCGCCAACCUGAGACCCACAAAUCGUCUUCCCCCCUUACAC-CC-- ...(((((............(((((((......))))))).....(((..(((...((.((((((....)))))).))....))).))).......)))))..............-..-- ( -28.00) >DroYak_CAF1 191 117 - 1 UCCAAUGAGUCGAACGGCCAGGUGGCUCAAAGCAGCCGCCAAAAAGUCAACAGAAAGCAAAUUGCCACCGCAAAUCGCCAACCUGAGACCCAGAAUUCGUCUUCCCCC-A--CACACAAA ......(((.((((.(((..(((((((......))))))).....................((((....))))...)))...(((.....)))..)))).))).....-.--........ ( -26.90) >consensus UCCGAUGAAUCGAACGGCCAGGUGGCUCAAAGCAGCCGCCAAAAAGUCAACAGAAAGCAAAUUGCCACCGCGAAUCGCCAACCUGAGACCCACAAAUCGUCUUCCCCCCA__CAC_CAAA ...(((((............((((((........)))))).....(((..(((...((...((((....))))...))....))).))).......)))))................... (-22.12 = -22.23 + 0.11)

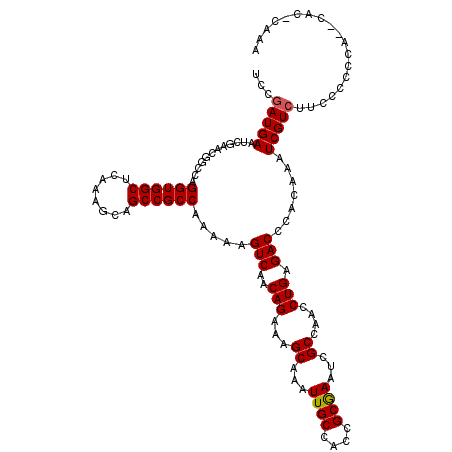
| Location | 16,132,193 – 16,132,313 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -40.37 |
| Consensus MFE | -35.14 |
| Energy contribution | -35.03 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16132193 120 + 23771897 UGGCUAUUCGCGGUGGCAAUUUGCUUUCUGUUGACUUUUUGGCGGCUGCUUUGUGCCACCUGGCCGUUCGAUUCAUCGGAUUGAUCGAUUCGAUCGCAUUGGAUCACAUAGAAUCGAUGG .(((((.....(((((((....((...((((..(....)..))))..))....))))))))))))..(((((((...(((((....)))))((((......)))).....)))))))... ( -43.60) >DroSim_CAF1 7420 120 + 1 UGGCGAUUCGCGGUGGCAAAUUGCUUUCUGUUGACUUUUUGGCGGCUGCUUUGAGCCACCUGGCCGUUUGAUUCAUCGGAUUGAUCGAUUCGAUCGCAUUGGAUCACAUAGAAUCGAUGG ...((((((((((((((..(..((...((((..(....)..))))..))..)..))))))..))....(((((((..(((((((.....)))))).)..)))))))....)))))).... ( -41.00) >DroYak_CAF1 228 120 + 1 UGGCGAUUUGCGGUGGCAAUUUGCUUUCUGUUGACUUUUUGGCGGCUGCUUUGAGCCACCUGGCCGUUCGACUCAUUGGAUUGAUCGAUUGGAUCGCAUUGGAUCACAUAGAAUCGAUUG ..(((((....((((((.....((...((((..(....)..))))..)).....))))))...(((.((((.(((......))))))).))))))))(((((.((.....)).))))).. ( -36.50) >consensus UGGCGAUUCGCGGUGGCAAUUUGCUUUCUGUUGACUUUUUGGCGGCUGCUUUGAGCCACCUGGCCGUUCGAUUCAUCGGAUUGAUCGAUUCGAUCGCAUUGGAUCACAUAGAAUCGAUGG .(((.(.....((((((.....((...((((..(....)..))))..)).....))))))).)))..((((((((((.((((((((.....))))).))).)))......)))))))... (-35.14 = -35.03 + -0.11)

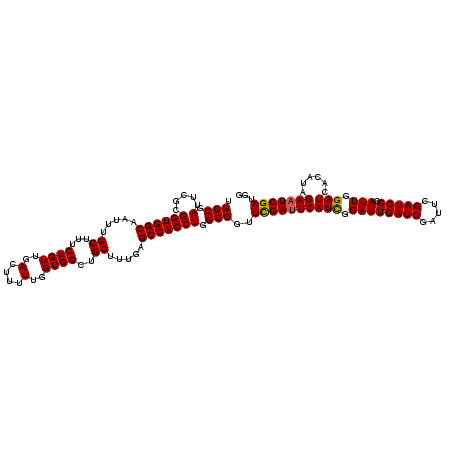

| Location | 16,132,193 – 16,132,313 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -29.61 |
| Energy contribution | -29.50 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16132193 120 - 23771897 CCAUCGAUUCUAUGUGAUCCAAUGCGAUCGAAUCGAUCAAUCCGAUGAAUCGAACGGCCAGGUGGCACAAAGCAGCCGCCAAAAAGUCAACAGAAAGCAAAUUGCCACCGCGAAUAGCCA ...(((((((....(((((......))))).((((.......)))))))))))..(((..((((((........)))))).....................((((....))))...))). ( -32.90) >DroSim_CAF1 7420 120 - 1 CCAUCGAUUCUAUGUGAUCCAAUGCGAUCGAAUCGAUCAAUCCGAUGAAUCAAACGGCCAGGUGGCUCAAAGCAGCCGCCAAAAAGUCAACAGAAAGCAAUUUGCCACCGCGAAUCGCCA .....(((((....(((((......))))).((((.......)))))))))....(((..(((((((......)))))))...................((((((....)))))).))). ( -31.80) >DroYak_CAF1 228 120 - 1 CAAUCGAUUCUAUGUGAUCCAAUGCGAUCCAAUCGAUCAAUCCAAUGAGUCGAACGGCCAGGUGGCUCAAAGCAGCCGCCAAAAAGUCAACAGAAAGCAAAUUGCCACCGCAAAUCGCCA ...(((((((..((.(((......((((...))))....)))))..)))))))..(((..(((((((......))))))).....................((((....))))...))). ( -31.80) >consensus CCAUCGAUUCUAUGUGAUCCAAUGCGAUCGAAUCGAUCAAUCCGAUGAAUCGAACGGCCAGGUGGCUCAAAGCAGCCGCCAAAAAGUCAACAGAAAGCAAAUUGCCACCGCGAAUCGCCA ...(((((((...((........))((((.....))))........)))))))..(((..((((((........)))))).....................((((....))))...))). (-29.61 = -29.50 + -0.11)

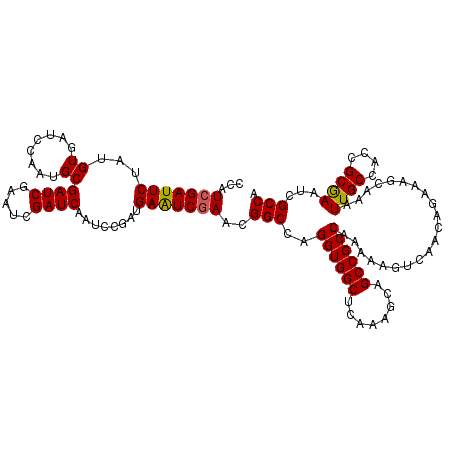
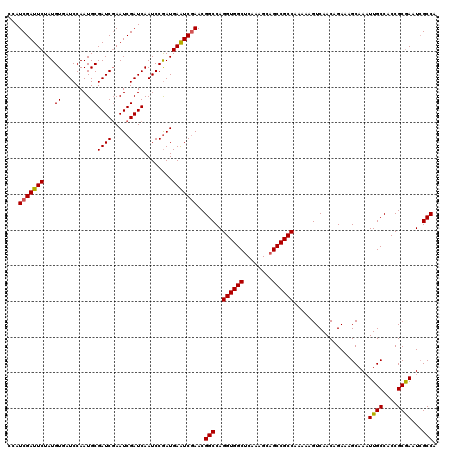
| Location | 16,132,233 – 16,132,353 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -43.65 |
| Energy contribution | -44.10 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16132233 120 + 23771897 GGCGGCUGCUUUGUGCCACCUGGCCGUUCGAUUCAUCGGAUUGAUCGAUUCGAUCGCAUUGGAUCACAUAGAAUCGAUGGGAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUG (((.((.((((((.(((....))).((.((((.(((....(((((((..((((((.((((((.((.....)).)))))).))))))..)))))))))))))).)))))))).))..))). ( -47.40) >DroSim_CAF1 7460 120 + 1 GGCGGCUGCUUUGAGCCACCUGGCCGUUUGAUUCAUCGGAUUGAUCGAUUCGAUCGCAUUGGAUCACAUAGAAUCGAUGGGAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUG (((.((.((((((.(((....))).((.((((.(((....(((((((..((((((.((((((.((.....)).)))))).))))))..)))))))))))))).)))))))).))..))). ( -46.50) >DroYak_CAF1 268 120 + 1 GGCGGCUGCUUUGAGCCACCUGGCCGUUCGACUCAUUGGAUUGAUCGAUUGGAUCGCAUUGGAUCACAUAGAAUCGAUUGGAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUG (((.((.((((((.(((....))).((.(((.(((((.((((((((((((.(((((...((.....))......))))).))))))).))))).)))))))).)))))))).))..))). ( -44.10) >consensus GGCGGCUGCUUUGAGCCACCUGGCCGUUCGAUUCAUCGGAUUGAUCGAUUCGAUCGCAUUGGAUCACAUAGAAUCGAUGGGAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUG (((.((.((((((.(((....))).((.(((.((((....(((((((..((((((.((((((.((.....)).)))))).))))))..)))))))))))))).)))))))).))..))). (-43.65 = -44.10 + 0.45)

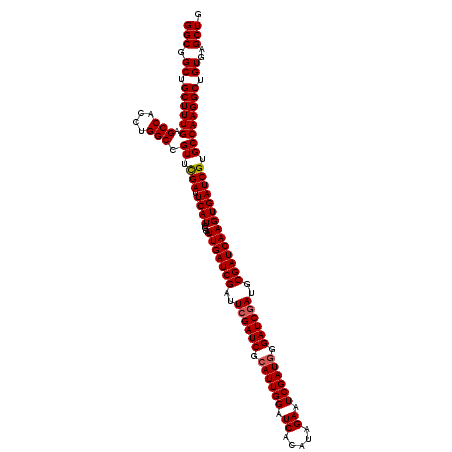
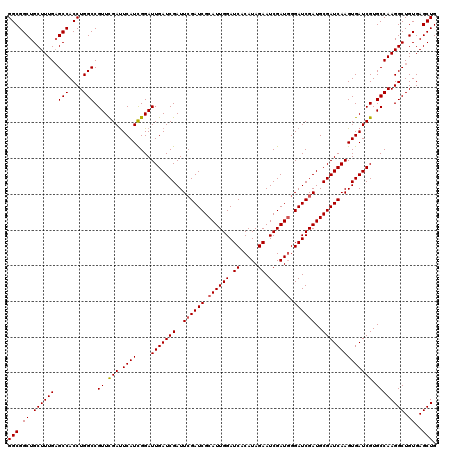
| Location | 16,132,233 – 16,132,353 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -36.65 |
| Energy contribution | -37.43 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16132233 120 - 23771897 CAGCUCACAGCCUUGGCACGAUCACUUGAUCGCAUCGAUCCCAUCGAUUCUAUGUGAUCCAAUGCGAUCGAAUCGAUCAAUCCGAUGAAUCGAACGGCCAGGUGGCACAAAGCAGCCGCC ..((.....))..((((.((.....((((((((((.((((.(((.......))).))))..)))))))))).(((((((......)).))))).))))))((((((........)))))) ( -40.90) >DroSim_CAF1 7460 120 - 1 CAGCUCACAGCCUUGGCACGAUCACUUGAUCGCAUCGAUCCCAUCGAUUCUAUGUGAUCCAAUGCGAUCGAAUCGAUCAAUCCGAUGAAUCAAACGGCCAGGUGGCUCAAAGCAGCCGCC ..((.....))..((((.(((((....)))))(((((.....((((((((.....((((......)))))))))))).....))))).........))))(((((((......))))))) ( -39.40) >DroYak_CAF1 268 120 - 1 CAGCUCACAGCCUUGGCACGAUCACUUGAUCGCAUCGAUCCAAUCGAUUCUAUGUGAUCCAAUGCGAUCCAAUCGAUCAAUCCAAUGAGUCGAACGGCCAGGUGGCUCAAAGCAGCCGCC ..((.....))..((((.((.......((((((((.((((((..........)).))))..))))))))...(((((((......))).)))).))))))(((((((......))))))) ( -38.10) >consensus CAGCUCACAGCCUUGGCACGAUCACUUGAUCGCAUCGAUCCCAUCGAUUCUAUGUGAUCCAAUGCGAUCGAAUCGAUCAAUCCGAUGAAUCGAACGGCCAGGUGGCUCAAAGCAGCCGCC ..((.....))..((((.((.....((((((((((.((((.(((.......))).))))..)))))))))).(((((((......)).))))).))))))((((((........)))))) (-36.65 = -37.43 + 0.78)



| Location | 16,132,273 – 16,132,393 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -32.50 |
| Energy contribution | -32.83 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16132273 120 + 23771897 UUGAUCGAUUCGAUCGCAUUGGAUCACAUAGAAUCGAUGGGAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUGUCUGUCUGCAUUUAUAAAUAAAUUGUGGCAAUUAAAUAAA (((((((..((((((.((((((.((.....)).)))))).))))))..))))))).......((((.((((........))))....(((((((....)))).))))))).......... ( -36.00) >DroSim_CAF1 7500 120 + 1 UUGAUCGAUUCGAUCGCAUUGGAUCACAUAGAAUCGAUGGGAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUGUCUGUCUGCAUUUAUAAAUAAAUUGUGGCAAUUAAAUAAA (((((((..((((((.((((((.((.....)).)))))).))))))..))))))).......((((.((((........))))....(((((((....)))).))))))).......... ( -36.00) >DroYak_CAF1 308 120 + 1 UUGAUCGAUUGGAUCGCAUUGGAUCACAUAGAAUCGAUUGGAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUGUCUGUCUGCAUUUAUAAAUAAAUUGUGGCAAUUAAAUAAA ..(((((....((((((((((.(((.((..........)))))))))))))))...))))).((((.((((........))))....(((((((....)))).))))))).......... ( -33.20) >consensus UUGAUCGAUUCGAUCGCAUUGGAUCACAUAGAAUCGAUGGGAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUGUCUGUCUGCAUUUAUAAAUAAAUUGUGGCAAUUAAAUAAA ..(((((....((((((((((.(((.(((.......))).)))))))))))))...))))).((((.((((........))))....(((((((....)))).))))))).......... (-32.50 = -32.83 + 0.33)

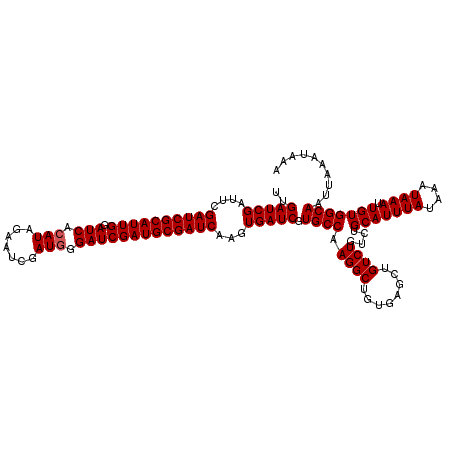

| Location | 16,132,273 – 16,132,393 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -26.33 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.939071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16132273 120 - 23771897 UUUAUUUAAUUGCCACAAUUUAUUUAUAAAUGCAGACAGACAGCUCACAGCCUUGGCACGAUCACUUGAUCGCAUCGAUCCCAUCGAUUCUAUGUGAUCCAAUGCGAUCGAAUCGAUCAA ..........(((((..(((((....)))))...........((.....))..))))).((((..((((((((((.((((.(((.......))).))))..))))))))))...)))).. ( -28.80) >DroSim_CAF1 7500 120 - 1 UUUAUUUAAUUGCCACAAUUUAUUUAUAAAUGCAGACAGACAGCUCACAGCCUUGGCACGAUCACUUGAUCGCAUCGAUCCCAUCGAUUCUAUGUGAUCCAAUGCGAUCGAAUCGAUCAA ..........(((((..(((((....)))))...........((.....))..))))).((((..((((((((((.((((.(((.......))).))))..))))))))))...)))).. ( -28.80) >DroYak_CAF1 308 120 - 1 UUUAUUUAAUUGCCACAAUUUAUUUAUAAAUGCAGACAGACAGCUCACAGCCUUGGCACGAUCACUUGAUCGCAUCGAUCCAAUCGAUUCUAUGUGAUCCAAUGCGAUCCAAUCGAUCAA ..........(((((..(((((....)))))...........((.....))..))))).((((..((((((((((.((((((..........)).))))..))))))).)))..)))).. ( -24.80) >consensus UUUAUUUAAUUGCCACAAUUUAUUUAUAAAUGCAGACAGACAGCUCACAGCCUUGGCACGAUCACUUGAUCGCAUCGAUCCCAUCGAUUCUAUGUGAUCCAAUGCGAUCGAAUCGAUCAA ..........(((((..(((((....)))))...........((.....))..))))).((((..((((((((((.((((.(((.......))).))))..))))))))))...)))).. (-26.33 = -27.00 + 0.67)

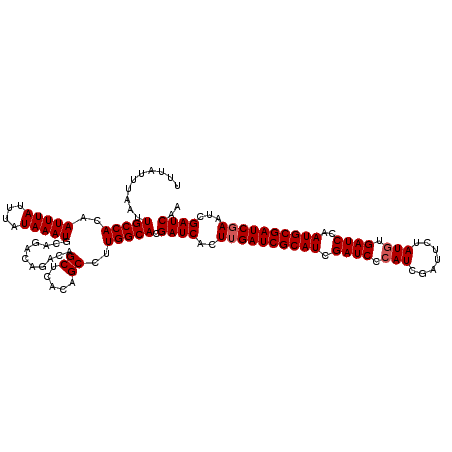
| Location | 16,132,313 – 16,132,433 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -28.96 |
| Energy contribution | -29.41 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16132313 120 + 23771897 GAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUGUCUGUCUGCAUUUAUAAAUAAAUUGUGGCAAUUAAAUAAAUAAAAUGGAUGCAUAUGUAUGCUUUUUUAAUUGUCAGCCA ....(..((((((....))))))..)..((((((..((.....))..)))...............(((((((((((..........(.((((....)))).)...)))))))))))))). ( -28.42) >DroSim_CAF1 7540 120 + 1 GAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUGUCUGUCUGCAUUUAUAAAUAAAUUGUGGCAAUUAAAUAAAUAAAAUGGUUGCAUAUGUAUGCAUUUUUAAUUGUCAGCCA ....(..((((((....))))))..)..((((((..((.....))..)))...............(((((((((((.............(((((....)))))..)))))))))))))). ( -29.36) >DroYak_CAF1 348 120 + 1 GAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUGUCUGUCUGCAUUUAUAAAUAAAUUGUGGCAAUUAAAUAAAUAAAAUGGAUGCAUAUGUAUGCAUUUUUAAUUGUCGGCCU ....(..((((((....))))))..).(((((((..((.....))..)).................((((((((((.......((((.((((....)))).))))))))))))))))))) ( -31.71) >consensus GAUCGAUGCGAUCAAGUGAUCGUGCCAAGGCUGUGAGCUGUCUGUCUGCAUUUAUAAAUAAAUUGUGGCAAUUAAAUAAAUAAAAUGGAUGCAUAUGUAUGCAUUUUUAAUUGUCAGCCA ....(..((((((....))))))..)..((((((..((.....))..)).................((((((((((.......((((.((((....)))).)))))))))))))))))). (-28.96 = -29.41 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:38 2006