
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,123,289 – 16,123,560 |
| Length | 271 |
| Max. P | 0.806421 |

| Location | 16,123,289 – 16,123,401 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.71 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -17.29 |
| Energy contribution | -15.97 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16123289 112 - 23771897 UCUUGGGUUUCUCAUACAGUGAGAGAAAAAAAAAGGGUAAUGCCAUUGGGUAAUCAAUUCAAUGGUACACUACUAUUUCAAUGCCAGUAUUU--------UAAAGGGGAAGUAGUUUAUG .(((...((((((((...))))))))......)))(((..(((((((((((.....))))))))))).)))((((((((....((.......--------....)).))))))))..... ( -26.50) >DroSec_CAF1 8976 108 - 1 UCUUGGGUUUCUUACACAGUGAGAAAA--AAAGAGGGGAAUGCCAUUAGGUAAACAAUCCAAUGGCACACUACUAU-UCAAUACCAGUAUUUAUGUAUUAUAAA---------AUGUAAA ..((((.((((((((...)))))))).--...(((.((..(((((((.((........)))))))))..))....)-))....))))..(((((((........---------))))))) ( -22.30) >DroSim_CAF1 8455 110 - 1 UCUUGGGUUUCUUACACAGUGAGAAAA-AAAAGAGGGGAAUGCCAUUAGGUAAUCAAUUCAAUGGCACACUACUAUUUCAAUGACAGUAUUUGUGUAUUAUAAA---------AUGUAAA ((((...((((((((...)))))))).-..)))).......((((((..((.....))..))))))((((((((.((.....)).))))...))))........---------....... ( -22.10) >consensus UCUUGGGUUUCUUACACAGUGAGAAAA_AAAAGAGGGGAAUGCCAUUAGGUAAUCAAUUCAAUGGCACACUACUAUUUCAAUGCCAGUAUUU_UGUAUUAUAAA_________AUGUAAA ((((...((((((((...))))))))......))))....(((((((.((........)))))))))..................................................... (-17.29 = -15.97 + -1.32)



| Location | 16,123,321 – 16,123,441 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -30.63 |
| Consensus MFE | -26.63 |
| Energy contribution | -25.87 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16123321 120 - 23771897 UGUCUCGACAAUCAGCAGACUCCCAUGACUUAGGCAACAUUCUUGGGUUUCUCAUACAGUGAGAGAAAAAAAAAGGGUAAUGCCAUUGGGUAAUCAAUUCAAUGGUACACUACUAUUUCA .(((((........).)))).((((.((....(....)..)).))))((((((((...)))))))).........((((.(((((((((((.....)))))))))))...))))...... ( -29.40) >DroSec_CAF1 9007 117 - 1 UGUCUCGACAAUCAGCAGACUCCCAUGACUUAGGCAACAUUCUUGGGUUUCUUACACAGUGAGAAAA--AAAGAGGGGAAUGCCAUUAGGUAAACAAUCCAAUGGCACACUACUAU-UCA .(((((........).))))((((........(....).(((((...((((((((...)))))))).--.))))))))).(((((((.((........))))))))).........-... ( -31.70) >DroSim_CAF1 8486 119 - 1 UGUCUCGACAAUCAGCAGACUCCCAUGACUUAGGCAACAUUCUUGGGUUUCUUACACAGUGAGAAAA-AAAAGAGGGGAAUGCCAUUAGGUAAUCAAUUCAAUGGCACACUACUAUUUCA .(((((........).))))((((........(....).(((((...((((((((...)))))))).-..))))))))).(((((((..((.....))..)))))))............. ( -30.80) >consensus UGUCUCGACAAUCAGCAGACUCCCAUGACUUAGGCAACAUUCUUGGGUUUCUUACACAGUGAGAAAA_AAAAGAGGGGAAUGCCAUUAGGUAAUCAAUUCAAUGGCACACUACUAUUUCA .(((((........).)))).(((........(....)..((((...((((((((...))))))))....)))))))...(((((((.((........)))))))))............. (-26.63 = -25.87 + -0.77)

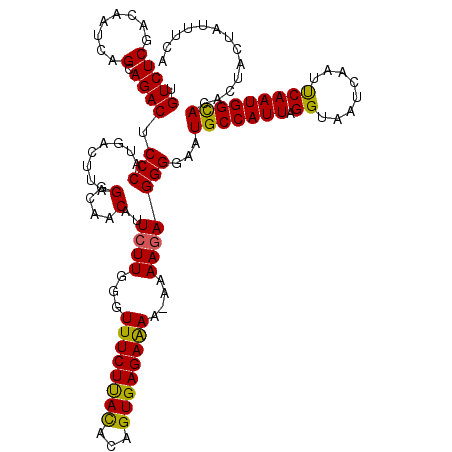

| Location | 16,123,441 – 16,123,560 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.84 |
| Mean single sequence MFE | -32.21 |
| Consensus MFE | -26.37 |
| Energy contribution | -27.37 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16123441 119 + 23771897 AAUUUCCAAGAAGCGAAAGAGUUUCUGUCGCUUUUUUUU-CCUUUUCUUUACUUUUCUUCAGCUAUCGCGCUAUUGUGUGGUUUAUUUAGUUGGGAAUGGGGUAGCGAUAUAUAUACGUA ........((((((......))))))((((((......(-(((.((((..(((...........(((((((....)))))))......)))..)))).)))).))))))........... ( -28.73) >DroSec_CAF1 9124 117 + 1 AAUUUCCAAGAAGCGAAAGAGUUUCUGUCGCUUUUUUUU-CGUUUUCUUUUCUUUUCUAGAGCUAUCGCGCUAUUGUGUGGUUUGUUUAGUUGGGGAUGGGGCAGCGAUAUAUAU--GUA .......(((((((((.((.....)).)))))))))..(-((((((((.(((((..(((((...(((((((....)))))))...)))))..))))).)))).))))).......--... ( -33.70) >DroSim_CAF1 8605 118 + 1 AAUUUCCAAGAAGCGAAAGAGUUUCUGUCGCUUUUUUUCCCCUUUUCUUUUCUUUUCUAGAGCUAUCGCGCUAUUGUGUGGUUUAUUUAGUUGGGGAUGGGGCAGCGAUAUAUAU--GUA ........((((((......))))))(((((((((..(((((..............(((((...(((((((....)))))))...)))))..)))))..))).))))))......--... ( -34.19) >consensus AAUUUCCAAGAAGCGAAAGAGUUUCUGUCGCUUUUUUUU_CCUUUUCUUUUCUUUUCUAGAGCUAUCGCGCUAUUGUGUGGUUUAUUUAGUUGGGGAUGGGGCAGCGAUAUAUAU__GUA ...(((((((((((((.((.....)).))))))))..............(((((..(((((...(((((((....)))))))...)))))..)))))))))).................. (-26.37 = -27.37 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:31 2006