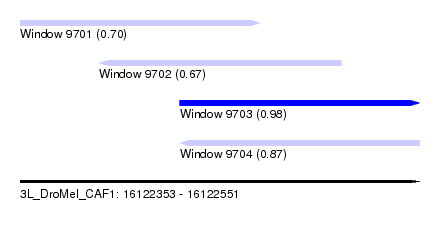
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,122,353 – 16,122,551 |
| Length | 198 |
| Max. P | 0.984524 |

| Location | 16,122,353 – 16,122,472 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -24.19 |
| Energy contribution | -23.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16122353 119 + 23771897 CACCUCCACCUUCACCUCC-CCCUGCCGCCUGUCUACGGGCAUUAUCAGCAGACAUAUAGUUUGGUUCCAUUUGUAAUCGCUCAUUGAUUCAGAAUUCCGAACUCACAAUGCCGAGUUUC ...............(((.-.......(((((....))))).......(((.......(((((((.....((((.(((((.....)))))))))...))))))).....))).))).... ( -23.80) >DroSec_CAF1 8056 119 + 1 CACCUCCACCCUCACCUCC-CCCUGCCGCCUGUCUACGGGCAUUAUCAGCAGACAUAUAGUUUGGUUCCAUUUGUAAUCGCUCAUUGAUUCAGAAUUCCGAACUCACAAUGCCGAGUUUC ..........(((......-.......(((((....))))).......(((.......(((((((.....((((.(((((.....)))))))))...))))))).....))).))).... ( -24.70) >DroSim_CAF1 7368 120 + 1 CACCUCCACCCUCACCUCCCCCCUGCCGCCUGUCUACGGGCAUUAUCAGCAGACAUAUAGUUUGGUUCCAUUUGUAAUCGCUCAUUGAUUCAGAAUUCCGAACUCACAAUGCCGAGUUUC ..........(((..............(((((....))))).......(((.......(((((((.....((((.(((((.....)))))))))...))))))).....))).))).... ( -24.70) >consensus CACCUCCACCCUCACCUCC_CCCUGCCGCCUGUCUACGGGCAUUAUCAGCAGACAUAUAGUUUGGUUCCAUUUGUAAUCGCUCAUUGAUUCAGAAUUCCGAACUCACAAUGCCGAGUUUC ..........(((..............(((((....))))).......(((.......(((((((.....((((.(((((.....)))))))))...))))))).....))).))).... (-24.19 = -23.97 + -0.22)


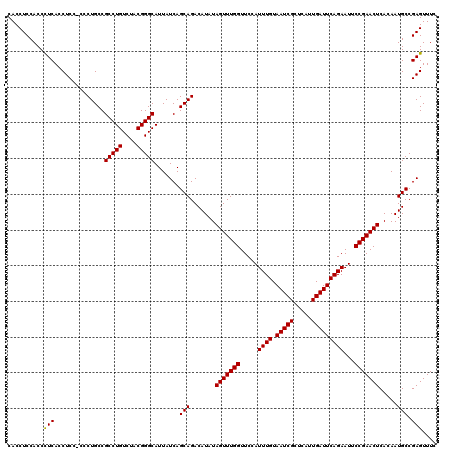
| Location | 16,122,392 – 16,122,512 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -30.03 |
| Energy contribution | -30.03 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16122392 120 - 23771897 AAUCGAUAUGGAAUCGCAAUGGAAACGCACACGUGCUGUGGAAACUCGGCAUUGUGAGUUCGGAAUUCUGAAUCAAUGAGCGAUUACAAAUGGAACCAAACUAUAUGUCUGCUGAUAAUG ..((.((.((.((((((...(....).((((.((((((.(....).)))))))))).((((((....))))))......)))))).)).)).))...........((((....))))... ( -34.40) >DroSec_CAF1 8095 120 - 1 AAUCGAUAUGGAAUCGCAAUGGAAACGCACACGUGCAUUGGAAACUCGGCAUUGUGAGUUCGGAAUUCUGAAUCAAUGAGCGAUUACAAAUGGAACCAAACUAUAUGUCUGCUGAUAAUG ..((.((.((.((((((...(....).((((.((((...(....)...)))))))).((((((....))))))......)))))).)).)).))...........((((....))))... ( -30.40) >DroSim_CAF1 7408 120 - 1 AAUCGAUAUGGAAUCGCAAUGGAAACGCACACGUGCAGUGGAAACUCGGCAUUGUGAGUUCGGAAUUCUGAAUCAAUGAGCGAUUACAAAUGGAACCAAACUAUAUGUCUGCUGAUAAUG ..((.((.((.((((((...(....).((((.((((.(.(....).).)))))))).((((((....))))))......)))))).)).)).))...........((((....))))... ( -30.50) >consensus AAUCGAUAUGGAAUCGCAAUGGAAACGCACACGUGCAGUGGAAACUCGGCAUUGUGAGUUCGGAAUUCUGAAUCAAUGAGCGAUUACAAAUGGAACCAAACUAUAUGUCUGCUGAUAAUG ..((.((.((.((((((...(....).((((.((((...(....)...)))))))).((((((....))))))......)))))).)).)).))...........((((....))))... (-30.03 = -30.03 + 0.00)

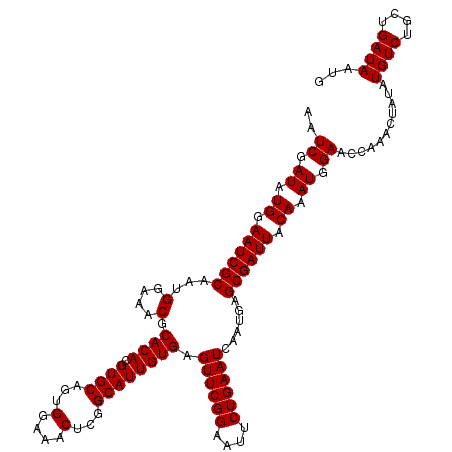

| Location | 16,122,432 – 16,122,551 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -35.66 |
| Consensus MFE | -33.21 |
| Energy contribution | -33.10 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16122432 119 + 23771897 CUCAUUGAUUCAGAAUUCCGAACUCACAAUGCCGAGUUUCCACAGCACGUGUGCGUUUCCAUUGCGAUUCCAUAUCGAUUCGCAAUGGAAUCAGACAUUA-AAACUGACACACAGUAUAU ...................((((((........)))))).........(((((...(((((((((((.((......)).)))))))))))((((......-...)))))))))....... ( -31.20) >DroSec_CAF1 8135 120 + 1 CUCAUUGAUUCAGAAUUCCGAACUCACAAUGCCGAGUUUCCAAUGCACGUGUGCGUUUCCAUUGCGAUUCCAUAUCGAUUCGCGAUGGAAUCGGACAUAAAAAACUGACACACGGUAUAU .....((((((.(....).))).)))..((((((.((.((........((((.((.(((((((((((.((......)).))))))))))).)).))))........)).)).)))))).. ( -36.49) >DroSim_CAF1 7448 120 + 1 CUCAUUGAUUCAGAAUUCCGAACUCACAAUGCCGAGUUUCCACUGCACGUGUGCGUUUCCAUUGCGAUUCCAUAUCGAUUCGCGAUGGAAGCGGACAUAAAAAACUGACACACAGUAUAU ...((((..((((......((((((........)))))).........((((.((((((((((((((.((......)).)))))))))))))).))))......))))....)))).... ( -39.30) >consensus CUCAUUGAUUCAGAAUUCCGAACUCACAAUGCCGAGUUUCCACUGCACGUGUGCGUUUCCAUUGCGAUUCCAUAUCGAUUCGCGAUGGAAUCGGACAUAAAAAACUGACACACAGUAUAU ...((((..((((......((((((........)))))).........((((.((.(((((((((((.((......)).))))))))))).)).))))......))))....)))).... (-33.21 = -33.10 + -0.11)


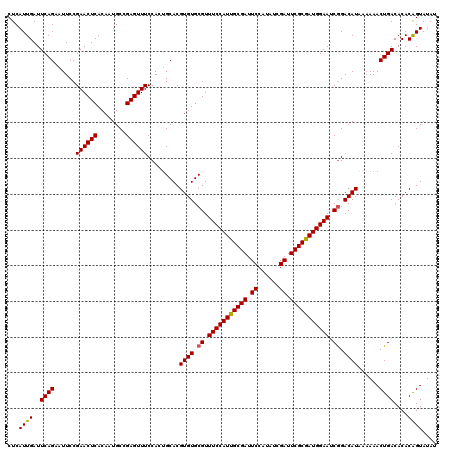
| Location | 16,122,432 – 16,122,551 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -33.22 |
| Energy contribution | -33.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16122432 119 - 23771897 AUAUACUGUGUGUCAGUUU-UAAUGUCUGAUUCCAUUGCGAAUCGAUAUGGAAUCGCAAUGGAAACGCACACGUGCUGUGGAAACUCGGCAUUGUGAGUUCGGAAUUCUGAAUCAAUGAG ......((((((((((...-......))))(((((((((((.((......)).))))))))))).)))))).((((((.(....).))))))..(((.(((((....))))))))..... ( -43.40) >DroSec_CAF1 8135 120 - 1 AUAUACCGUGUGUCAGUUUUUUAUGUCCGAUUCCAUCGCGAAUCGAUAUGGAAUCGCAAUGGAAACGCACACGUGCAUUGGAAACUCGGCAUUGUGAGUUCGGAAUUCUGAAUCAAUGAG ......(((...((((.........(((((((((((.((((.((......)).)))).))))))((.((((.((((...(....)...)))))))).)))))))...))))....))).. ( -36.80) >DroSim_CAF1 7448 120 - 1 AUAUACUGUGUGUCAGUUUUUUAUGUCCGCUUCCAUCGCGAAUCGAUAUGGAAUCGCAAUGGAAACGCACACGUGCAGUGGAAACUCGGCAUUGUGAGUUCGGAAUUCUGAAUCAAUGAG ...(((...(((((((((.......(((((((((((.((((.((......)).)))).))))))..(((....))).))))))))).))))).))).((((((....))))))....... ( -35.81) >consensus AUAUACUGUGUGUCAGUUUUUUAUGUCCGAUUCCAUCGCGAAUCGAUAUGGAAUCGCAAUGGAAACGCACACGUGCAGUGGAAACUCGGCAUUGUGAGUUCGGAAUUCUGAAUCAAUGAG ......(((((((((........))...(.((((((.((((.((......)).)))).)))))).))))))))(((...(....)...)))(..(..((((((....))))))..)..). (-33.22 = -33.00 + -0.22)

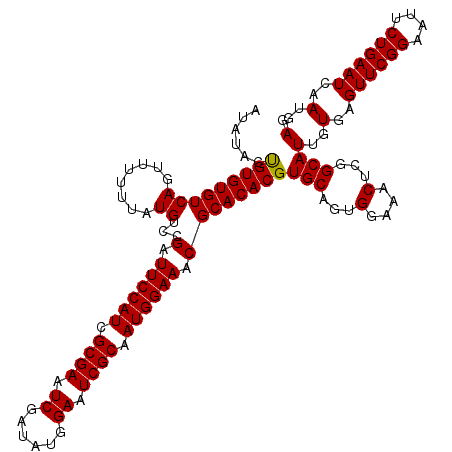
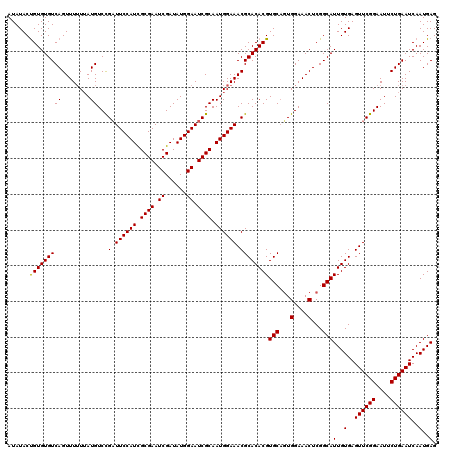
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:28 2006