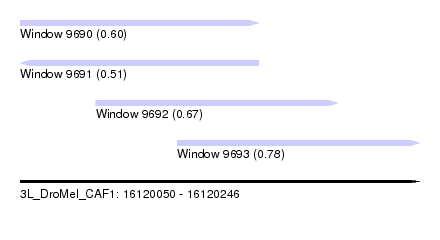
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,120,050 – 16,120,246 |
| Length | 196 |
| Max. P | 0.776700 |

| Location | 16,120,050 – 16,120,167 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -33.47 |
| Energy contribution | -33.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16120050 117 + 23771897 GUGUACAGCACU---GUCUGCGCUCCAGUUUCUUCGGCUCGGAUGAUUUUCCAGCUGAGAAUGAUAUAUGGCAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGG .....(((((((---(((.....((....((((.(((((.(((......)))))))))))).)).....))))))..))))(((((((..((((((.....))))))..))))))).... ( -42.30) >DroSec_CAF1 5772 117 + 1 GUGUACAGCACU---GUCUGCGCUCCAGUUUCUUCGGCUCGGAUGAUUUUCCAGCUCGGAAUGAUAUAUGACAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGG .....(((((((---(((.........(((((...((((.(((......))))))).))))).......))))))..))))(((((((..((((((.....))))))..))))))).... ( -39.69) >DroSim_CAF1 5125 120 + 1 GUGUACAGCACUGCUGUCUGCGCUCCAGUUUCUUCGGCUCGGAUGAUUUUCCAGCUCGGAAUGAUAUAUGGCAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGG .....((((((((((((.((((.(((.(......)((((.(((......))))))).))).)).)).))))))))..))))(((((((..((((((.....))))))..))))))).... ( -41.60) >consensus GUGUACAGCACU___GUCUGCGCUCCAGUUUCUUCGGCUCGGAUGAUUUUCCAGCUCGGAAUGAUAUAUGGCAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGG .....((((......(....)((((((...(((((((((.(((......)))))))..))).))....))).)))..))))(((((((..((((((.....))))))..))))))).... (-33.47 = -33.80 + 0.33)

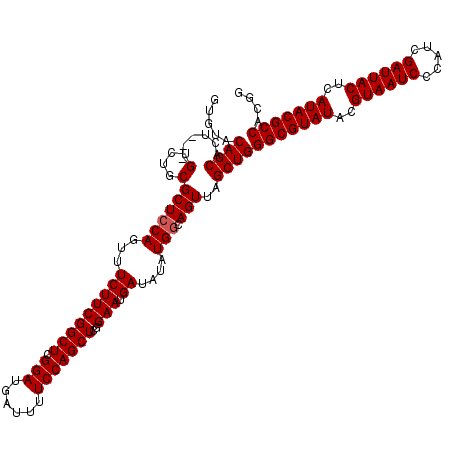

| Location | 16,120,050 – 16,120,167 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -32.70 |
| Energy contribution | -32.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16120050 117 - 23771897 CCGUGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACUGCCAUAUAUCAUUCUCAGCUGGAAAAUCAUCCGAGCCGAAGAAACUGGAGCGCAGAC---AGUGCUGUACAC ....((((((((.((((((.....)))))).))))))))((((..((((..........(((((.((((((......))).))).).)))).(((.....))).)---)))))))..... ( -36.40) >DroSec_CAF1 5772 117 - 1 CCGUGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACUGUCAUAUAUCAUUCCGAGCUGGAAAAUCAUCCGAGCCGAAGAAACUGGAGCGCAGAC---AGUGCUGUACAC ....((((((((.((((((.....)))))).))))))))((((..((((((.....((.(((((.((((((......))).)))))..)))....)).....)))---)))))))..... ( -38.70) >DroSim_CAF1 5125 120 - 1 CCGUGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACUGCCAUAUAUCAUUCCGAGCUGGAAAAUCAUCCGAGCCGAAGAAACUGGAGCGCAGACAGCAGUGCUGUACAC ....((((((((.((((((.....)))))).))))))))((((..(((((............((.((((((......))).)))))......(((.....)))...)))))))))..... ( -39.00) >consensus CCGUGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACUGCCAUAUAUCAUUCCGAGCUGGAAAAUCAUCCGAGCCGAAGAAACUGGAGCGCAGAC___AGUGCUGUACAC (((.((((((((.((((((.....)))))).)))))))))................((.(((...((((((......))).))).)))))....))(((((........)))))...... (-32.70 = -32.70 + -0.00)

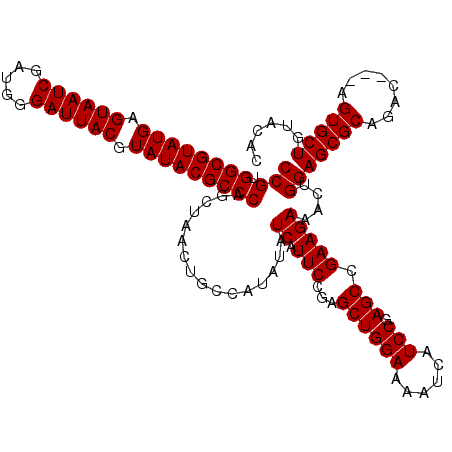

| Location | 16,120,087 – 16,120,206 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -36.43 |
| Energy contribution | -37.77 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16120087 119 + 23771897 GGAUGAUUUUCCAGCUGAGAAUGAUAUAUGGCAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGAGCCGUGGAAUUGCGUUGUU-GCGUUACAGCAA (((......))).((((..((((..(((..((((((.....(((((((..((((((.....))))))..)))))))((((.((...)).))))..))))))..))).-.)))).)))).. ( -39.90) >DroSec_CAF1 5809 120 + 1 GGAUGAUUUUCCAGCUCGGAAUGAUAUAUGACAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGCGCCGGCGAAUUGCGUUGUUGGCGUUACAGCAA (((......))).(((((.............(((....)))(((((((..((((((.....))))))..))))))).)))))...(((((((((((......)))))))))))....... ( -42.50) >DroSim_CAF1 5165 120 + 1 GGAUGAUUUUCCAGCUCGGAAUGAUAUAUGGCAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGCGCCGGCGAAUUGCGUUGUUGGCGUUACAGCAA (((......))).(((.(...((((...((.(((....)))(((((((..((((((.....))))))..))))))).)).)))).(((((((((((......))))))))))).)))).. ( -42.40) >consensus GGAUGAUUUUCCAGCUCGGAAUGAUAUAUGGCAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGCGCCGGCGAAUUGCGUUGUUGGCGUUACAGCAA (((......))).(((.....((((...((.(((....)))(((((((..((((((.....))))))..))))))).)).)))).(((((((((((......)))))))))))..))).. (-36.43 = -37.77 + 1.33)


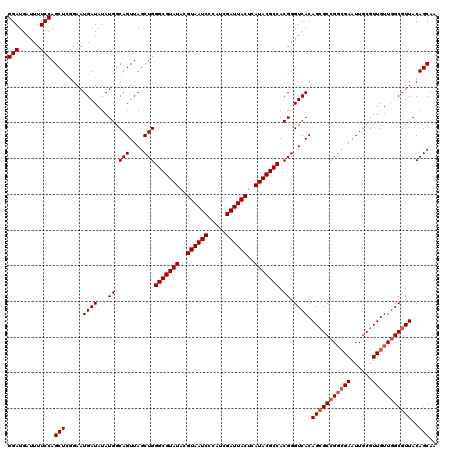
| Location | 16,120,127 – 16,120,246 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -37.33 |
| Consensus MFE | -32.33 |
| Energy contribution | -33.00 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16120127 119 + 23771897 GGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGAGCCGUGGAAUUGCGUUGUU-GCGUUACAGCAAUUGUCUGCAGAUAAGCAAAACCAUUUCAAUUAAUAUCUUC .(((((((..((((((.....))))))..)))))))((((.((...)).))))((..(((((((((.-.....)))))..(((((....)))))))))..)).................. ( -36.40) >DroSec_CAF1 5849 120 + 1 GGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGCGCCGGCGAAUUGCGUUGUUGGCGUUACAGCAAUUGUCUGCAGAUAAGCAAAGCCAUUUCAAUUAAUUUCUUC .(((((((..((((((.....))))))..)))))))...(((...(((((((((((......)))))))))))...((..(((((....)))))))...))).................. ( -37.80) >DroSim_CAF1 5205 120 + 1 GGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGCGCCGGCGAAUUGCGUUGUUGGCGUUACAGCAAUUGUCUGCAGAUAAGCAAAGCCAUUUCAAUUAAUUUCUUC .(((((((..((((((.....))))))..)))))))...(((...(((((((((((......)))))))))))...((..(((((....)))))))...))).................. ( -37.80) >consensus GGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGCGCCGGCGAAUUGCGUUGUUGGCGUUACAGCAAUUGUCUGCAGAUAAGCAAAGCCAUUUCAAUUAAUUUCUUC .(((((((..((((((.....))))))..)))))))...(((......)))(((...(((((((((.......)))))..(((((....))))))))).))).................. (-32.33 = -33.00 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:18 2006