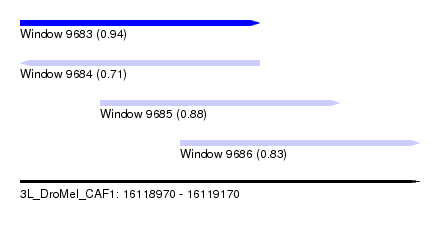
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,118,970 – 16,119,170 |
| Length | 200 |
| Max. P | 0.935682 |

| Location | 16,118,970 – 16,119,090 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -32.87 |
| Consensus MFE | -28.79 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16118970 120 + 23771897 AGAUUUGUUCAUCAUAAUUUGCAAACGCAUUUAAUUAAUGGGAAAUCACGUGGUGAACAUUGUGCAAACACCUCGUAUGCGCAAUAAUUCAGCUACGUUACGUAUACGCAGCGUAUCGCA ......(((((((((....(((....))).........(((....))).)))))))))((((((((.((.....)).))))))))......((((((((.((....)).))))))..)). ( -31.50) >DroSec_CAF1 4722 120 + 1 AGAUUUGUUCAUCAUAAUUCGCAAACGCAUUUAAUUAAUGGGAAAUCACGUGGUGAACAUUGUGCAAACACCUCGUAUGCGCAAUAAUUCAGCUGCGUUACGUAUACGCAGCGUAUCGCA .(((..(((((((((.....((....))..........(((....))).)))))))))((((((((.((.....)).))))))))......(((((((.......)))))))..)))... ( -35.10) >DroSim_CAF1 4081 109 + 1 -----------UCAUAAUUCGCAAACGCAUUUAAUUAAUGGGAAAUCACGUGGCGAACAUUGUGCAAACACCUCGUAUGCGCAAUAAUUCAGCUGCGUUACGUAUACGCAGCGUAUCGCA -----------..............(.((((.....)))).)..........((((((((((((((.((.....)).))))))))......(((((((.......))))))))).)))). ( -32.00) >consensus AGAUUUGUUCAUCAUAAUUCGCAAACGCAUUUAAUUAAUGGGAAAUCACGUGGUGAACAUUGUGCAAACACCUCGUAUGCGCAAUAAUUCAGCUGCGUUACGUAUACGCAGCGUAUCGCA .........................(.((((.....)))).)..........((((((((((((((.((.....)).))))))))......(((((((.......))))))))).)))). (-28.79 = -28.90 + 0.11)

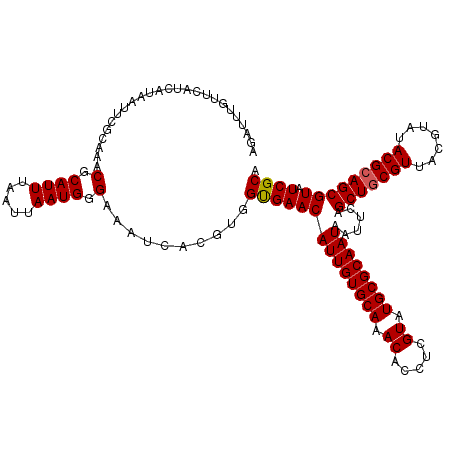

| Location | 16,118,970 – 16,119,090 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -29.99 |
| Energy contribution | -29.33 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.30 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16118970 120 - 23771897 UGCGAUACGCUGCGUAUACGUAACGUAGCUGAAUUAUUGCGCAUACGAGGUGUUUGCACAAUGUUCACCACGUGAUUUCCCAUUAAUUAAAUGCGUUUGCAAAUUAUGAUGAACAAAUCU (((((...(((((((.......))))))).........(((((((((.((((...((.....)).)))).)))((((.......))))..)))))))))))......(((......))). ( -29.30) >DroSec_CAF1 4722 120 - 1 UGCGAUACGCUGCGUAUACGUAACGCAGCUGAAUUAUUGCGCAUACGAGGUGUUUGCACAAUGUUCACCACGUGAUUUCCCAUUAAUUAAAUGCGUUUGCGAAUUAUGAUGAACAAAUCU .((((...(((((((.......))))))).........(((((((((.((((...((.....)).)))).)))((((.......))))..)))))))))).......(((......))). ( -30.60) >DroSim_CAF1 4081 109 - 1 UGCGAUACGCUGCGUAUACGUAACGCAGCUGAAUUAUUGCGCAUACGAGGUGUUUGCACAAUGUUCGCCACGUGAUUUCCCAUUAAUUAAAUGCGUUUGCGAAUUAUGA----------- .((((...(((((((.......))))))).........(((((((((.((((...((.....)).)))).)))((((.......))))..)))))))))).........----------- ( -29.50) >consensus UGCGAUACGCUGCGUAUACGUAACGCAGCUGAAUUAUUGCGCAUACGAGGUGUUUGCACAAUGUUCACCACGUGAUUUCCCAUUAAUUAAAUGCGUUUGCGAAUUAUGAUGAACAAAUCU (((((...(((((((.......))))))).........(((((((((.((((...((.....)).)))).)))((((.......))))..)))))))))))................... (-29.99 = -29.33 + -0.66)



| Location | 16,119,010 – 16,119,130 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -34.73 |
| Energy contribution | -35.73 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16119010 120 + 23771897 GGAAAUCACGUGGUGAACAUUGUGCAAACACCUCGUAUGCGCAAUAAUUCAGCUACGUUACGUAUACGCAGCGUAUCGCAUUGGGUGUGUUUGCUAUUUCAUCAUCUUUCUCGUAGCGAC (((((....((((((((.((...((((((((((((.((((((.........))((((((.((....)).))))))..)))))))).)))))))).)))))))))).)))))......... ( -35.80) >DroSec_CAF1 4762 120 + 1 GGAAAUCACGUGGUGAACAUUGUGCAAACACCUCGUAUGCGCAAUAAUUCAGCUGCGUUACGUAUACGCAGCGUAUCGCAUUUGGUGUGUUUGCUAUUUCAUCAUCUUUCUCGUAGCGAC (((((....((((((((.((...((((((((.(((.(((((..(((.....(((((((.......))))))).)))))))).))).)))))))).)))))))))).)))))......... ( -39.20) >DroSim_CAF1 4110 120 + 1 GGAAAUCACGUGGCGAACAUUGUGCAAACACCUCGUAUGCGCAAUAAUUCAGCUGCGUUACGUAUACGCAGCGUAUCGCAUUUGGUGUGUUUGCUAUUUCAUCAUCUUUCUCGUAGCGAC ........(((.((((((...))((((((((.(((.(((((..(((.....(((((((.......))))))).)))))))).))).))))))))................)))).))).. ( -36.10) >consensus GGAAAUCACGUGGUGAACAUUGUGCAAACACCUCGUAUGCGCAAUAAUUCAGCUGCGUUACGUAUACGCAGCGUAUCGCAUUUGGUGUGUUUGCUAUUUCAUCAUCUUUCUCGUAGCGAC (((((....((((((((.((...((((((((.(((.(((((..(((.....(((((((.......))))))).)))))))).))).)))))))).)))))))))).)))))......... (-34.73 = -35.73 + 1.00)


| Location | 16,119,050 – 16,119,170 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -33.05 |
| Energy contribution | -33.50 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16119050 120 + 23771897 GCAAUAAUUCAGCUACGUUACGUAUACGCAGCGUAUCGCAUUGGGUGUGUUUGCUAUUUCAUCAUCUUUCUCGUAGCGACGACUUGAUAAUAGAGUAGGCAAAUUCUAGAUGCAUAAGGA ...........(.((((((.((....)).)))))).)(((((.((...((((((((((..((.(((....((((....))))...))).))..)))).)))))).)).)))))....... ( -28.20) >DroSec_CAF1 4802 120 + 1 GCAAUAAUUCAGCUGCGUUACGUAUACGCAGCGUAUCGCAUUUGGUGUGUUUGCUAUUUCAUCAUCUUUCUCGUAGCGACGACUUGAUAAUAGAGCAGGCAAAUUCUAGAUGCAUUAGGA ...........(((((((.......))))))).....((((((((..((((((((.....((((......((((....))))..)))).....))))))))....))))))))....... ( -37.20) >DroSim_CAF1 4150 120 + 1 GCAAUAAUUCAGCUGCGUUACGUAUACGCAGCGUAUCGCAUUUGGUGUGUUUGCUAUUUCAUCAUCUUUCUCGUAGCGACGACUUGAUAAUAGAGUAGGCAAAUUCUAGAUGCAUUGGGA .((((......(((((((.......))))))).....((((((((...((((((((((..((.(((....((((....))))...))).))..)))).)))))).))))))))))))... ( -38.00) >consensus GCAAUAAUUCAGCUGCGUUACGUAUACGCAGCGUAUCGCAUUUGGUGUGUUUGCUAUUUCAUCAUCUUUCUCGUAGCGACGACUUGAUAAUAGAGUAGGCAAAUUCUAGAUGCAUUAGGA ...........(((((((.......))))))).....((((((((..((((((((.....((((......((((....))))..)))).....))))))))....))))))))....... (-33.05 = -33.50 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:01 2006