
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,117,521 – 16,117,748 |
| Length | 227 |
| Max. P | 0.889768 |

| Location | 16,117,521 – 16,117,628 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.48 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -20.34 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16117521 107 + 23771897 GCUGGCGCACAACAAAAGUUAUUAAUUCAAUGAUUAUGAUCAACGCGCUAAUGACAGUCAAGUGCAGCACUUGAUC-------------CGCAUAGAGAAAAGUGGCGAAAAAAAAAAAA ((((((((..(((....)))..........((((....))))..))))).(((...((((((((...)))))))).-------------..)))..........)))............. ( -22.30) >DroSec_CAF1 3340 116 + 1 GCUGGCACACAACAAAAGUUAUUAAUUCAAUGAUUAUGAUCAACGCGCUAAUGACAGUCAAGUGCAGCACUUGAUCCCACCCACCUACUCGCAUAGAGAAAAGUGCCGAAAAA----AAA ..((((((.........(((((((......((((....))))......))))))).((((((((...))))))))............(((.....)))....)))))).....----... ( -25.90) >DroSim_CAF1 2678 116 + 1 GCUGGUGCACAACAAAAGUUAUUAAUUCAAUGAUUAUGAUCAACGCGCUAAUGACAGUCAAGUGCAGCACUUGAUCCCACCCACCUCCUCGCAUAGAGAAAAGUGCCGAAAAA----AAA ..(((..(.........(((((((......((((....))))......))))))).((((((((...))))))))............(((.....)))....)..))).....----... ( -22.90) >consensus GCUGGCGCACAACAAAAGUUAUUAAUUCAAUGAUUAUGAUCAACGCGCUAAUGACAGUCAAGUGCAGCACUUGAUCCCACCCACCU_CUCGCAUAGAGAAAAGUGCCGAAAAA____AAA ..((((((.........(((((((......((((....))))......))))))).((((((((...))))))))...........................))))))............ (-20.34 = -20.23 + -0.11)

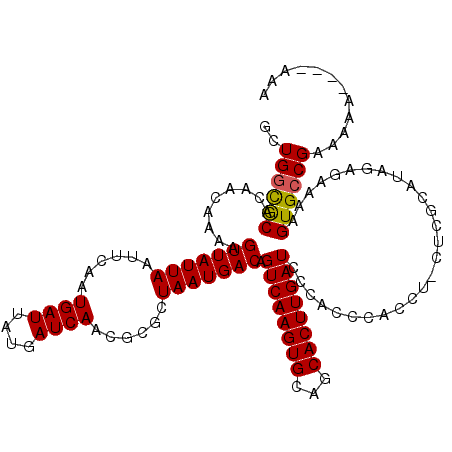
| Location | 16,117,597 – 16,117,708 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.73 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -33.85 |
| Energy contribution | -33.97 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16117597 111 + 23771897 ---------CGCAUAGAGAAAAGUGGCGAAAAAAAAAAAAGAUUCGGAGGCAAUGCCAUUGACACCUUGAUUGGAUGUCAAGGUUGUGCGCGAGGAUCAUCGCAUUCUCGCCGGGUGCUU ---------.((((.........((((((....................(((..(((.((((((((......)).)))))))))..)))((((......))))....)))))).)))).. ( -34.00) >DroSec_CAF1 3420 116 + 1 CCACCUACUCGCAUAGAGAAAAGUGCCGAAAAA----AAACAUUCGGAGGCAAUGCCAUUGACACCUUGAUUGGAUGUCAAGGUUGUGCGCGAGGAUCAUCGCAUUCUCGCCGGGUGCUU .(((((.(((((((........))))((((...----.....)))))))(((..(((.((((((((......)).)))))))))..)))((((((((......)))))))).)))))... ( -41.00) >DroSim_CAF1 2758 116 + 1 CCACCUCCUCGCAUAGAGAAAAGUGCCGAAAAA----AAACAUUCGGAGGCGAUGCCAUUGACACCUUGAUUGGAUGUCAAGGUUGUGCGCGAGGAUCAUCGCAUUCUCGCCGGGUGCUU .....(((((((.............(((((...----.....)))))..(((..(((.((((((((......)).)))))))))..)))))))))).....((((((.....)))))).. ( -40.90) >consensus CCACCU_CUCGCAUAGAGAAAAGUGCCGAAAAA____AAACAUUCGGAGGCAAUGCCAUUGACACCUUGAUUGGAUGUCAAGGUUGUGCGCGAGGAUCAUCGCAUUCUCGCCGGGUGCUU ..........((((.(((((.....(((((............)))))..(((..(((.((((((((......)).)))))))))..)))((((......)))).))))).....)))).. (-33.85 = -33.97 + 0.11)



| Location | 16,117,628 – 16,117,748 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -38.29 |
| Energy contribution | -38.07 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16117628 120 + 23771897 GAUUCGGAGGCAAUGCCAUUGACACCUUGAUUGGAUGUCAAGGUUGUGCGCGAGGAUCAUCGCAUUCUCGCCGGGUGCUUAAACUAACUAAGGCCAUUAUCAUAAGCUCCCAGCAAUCAU ((((..((((((..(((.((((((((......)).)))))))))..)))((((......))))...)))((.(((.(((((.....................))))).))).)))))).. ( -39.70) >DroSec_CAF1 3456 120 + 1 CAUUCGGAGGCAAUGCCAUUGACACCUUGAUUGGAUGUCAAGGUUGUGCGCGAGGAUCAUCGCAUUCUCGCCGGGUGCUUAAACUAACUAAGGCCAUUAUCAUAAGCUCCCAGCAAUCAU ......((((((..(((.((((((((......)).)))))))))..)))((((......))))...)))((.(((.(((((.....................))))).))).))...... ( -38.30) >DroSim_CAF1 2794 120 + 1 CAUUCGGAGGCGAUGCCAUUGACACCUUGAUUGGAUGUCAAGGUUGUGCGCGAGGAUCAUCGCAUUCUCGCCGGGUGCUUAAACUAACUGAGGCCAUUAUCAUAAGCUCCCAGCAAUCAU ......((((((..(((.((((((((......)).)))))))))..)))((((......))))...)))((.(((.(((((.......(((........)))))))).))).))...... ( -39.01) >consensus CAUUCGGAGGCAAUGCCAUUGACACCUUGAUUGGAUGUCAAGGUUGUGCGCGAGGAUCAUCGCAUUCUCGCCGGGUGCUUAAACUAACUAAGGCCAUUAUCAUAAGCUCCCAGCAAUCAU ......((((((..(((.((((((((......)).)))))))))..)))((((......))))...)))((.(((.(((((.....................))))).))).))...... (-38.29 = -38.07 + -0.22)



| Location | 16,117,628 – 16,117,748 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -38.70 |
| Energy contribution | -39.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16117628 120 - 23771897 AUGAUUGCUGGGAGCUUAUGAUAAUGGCCUUAGUUAGUUUAAGCACCCGGCGAGAAUGCGAUGAUCCUCGCGCACAACCUUGACAUCCAAUCAAGGUGUCAAUGGCAUUGCCUCCGAAUC ..((((((((((.(((((...((((.......))))...))))).))))))(((...((((......))))(((...((((((((((.......)))))))).))...))))))..)))) ( -40.20) >DroSec_CAF1 3456 120 - 1 AUGAUUGCUGGGAGCUUAUGAUAAUGGCCUUAGUUAGUUUAAGCACCCGGCGAGAAUGCGAUGAUCCUCGCGCACAACCUUGACAUCCAAUCAAGGUGUCAAUGGCAUUGCCUCCGAAUG ......((((((.(((((...((((.......))))...))))).))))))(((...((((......))))(((...((((((((((.......)))))))).))...))))))...... ( -39.80) >DroSim_CAF1 2794 120 - 1 AUGAUUGCUGGGAGCUUAUGAUAAUGGCCUCAGUUAGUUUAAGCACCCGGCGAGAAUGCGAUGAUCCUCGCGCACAACCUUGACAUCCAAUCAAGGUGUCAAUGGCAUCGCCUCCGAAUG ((..((((((((.(((((...((((.......))))...))))).))))))))..))((((((..((....(.(((.((((((.......))))))))))...))))))))......... ( -38.20) >consensus AUGAUUGCUGGGAGCUUAUGAUAAUGGCCUUAGUUAGUUUAAGCACCCGGCGAGAAUGCGAUGAUCCUCGCGCACAACCUUGACAUCCAAUCAAGGUGUCAAUGGCAUUGCCUCCGAAUG ......((((((.(((((...((((.......))))...))))).))))))(((...((((......))))(((...((((((((((.......)))))))).))...))))))...... (-38.70 = -39.03 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:56 2006