
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,078,923 – 16,079,233 |
| Length | 310 |
| Max. P | 0.994545 |

| Location | 16,078,923 – 16,079,043 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.07 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -31.48 |
| Energy contribution | -30.96 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16078923 120 - 23771897 UGGACGUUGCCAGAUAGCGUGCGAAAAACACGUAACCAACGAUCUGGCAAUGCCGCUCGUUUAUGCGGGUGGCGAGUGCGCAAUAUUCGUGAAUGCACUAAAUUUAAAAAAAAGUUCAAC (((((((((((((((.(((((.......)))))........))))))))))(((((((((....))))))))).((((((..((....))...))))))..............))))).. ( -43.60) >DroSec_CAF1 822 119 - 1 UGGACGUUGCCAGAUAGCGUGCGAAAAACACGUAACCAACGAUCUGGCAAUGCCGCGCGUUUAUGCGGGUGGCGAGUGUGCAAUAUUCGUGAAUGCACUAAAUUUAAA-AAAAGUUCAAC (((((((((((((((.(((((.......)))))........))))))))))(((((.(((....))).)))))....(((((...........)))))..........-....))))).. ( -41.10) >DroSim_CAF1 445 119 - 1 UGGACGUUGCCAGAUAGCGAGCGAAAAACACGUAACCAACGAUCUGGCAAUGCCGCGCGUUUAUGCGGGUGGCGACUGUGCAAUAUUUGUGAAUGCACUAAAUUUAAA-AAAAGUUCAAC (((((((((((((((.(....)........(((.....)))))))))))))(((((.(((....))).)))))....(((((...........)))))..........-....))))).. ( -38.20) >DroEre_CAF1 1225 119 - 1 CGGGCGUUGCCAGAUACCGUGCGAAAAACACGUAACCAAC-AUCUGGCAAUGCUGCGCGUUUAUGCGGGAGGCGAGUGCGCAAUCUUCGUGAAUGCACCAAGUUUAAAAAUAUGUUCAAC .((((((((((((((....((((.......))))......-))))))))))))...(((((((((.(((..(((....)))..))).))))))))).))..................... ( -41.80) >DroYak_CAF1 417 120 - 1 CGAGCGUUGCCAGAUAUCUUGCGAAAAACACGUAACCAACGACCUGGCAAUGCUGCGCAUUUAUGCGGGUGGAGAGUGCUCGAUCUUCAUGAAUGCACCAAGUUUAAAAAAUAGUUGAAA ..(((((((((((.....(((((.......)))))........)))))))))))..(((((((((.((((.(((....))).)))).)))))))))........................ ( -39.92) >consensus UGGACGUUGCCAGAUAGCGUGCGAAAAACACGUAACCAACGAUCUGGCAAUGCCGCGCGUUUAUGCGGGUGGCGAGUGCGCAAUAUUCGUGAAUGCACUAAAUUUAAAAAAAAGUUCAAC (((.(((((((((((....((((.......)))).......)))))))))))))).(((((((((...((.(((....))).))...)))))))))........................ (-31.48 = -30.96 + -0.52)

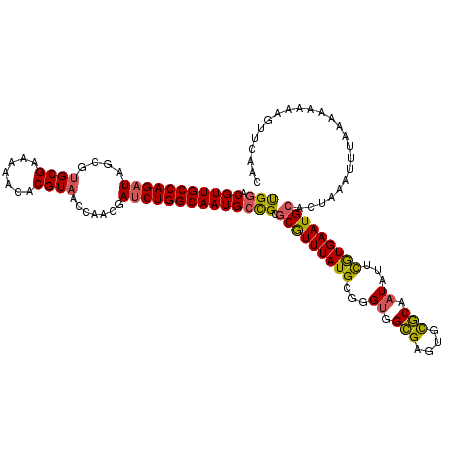
| Location | 16,078,963 – 16,079,083 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -41.34 |
| Consensus MFE | -32.78 |
| Energy contribution | -33.30 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16078963 120 - 23771897 AUUUUCACGAAUGCGCGUGCAAAGCGAAAAUCGAGGCUGGUGGACGUUGCCAGAUAGCGUGCGAAAAACACGUAACCAACGAUCUGGCAAUGCCGCUCGUUUAUGCGGGUGGCGAGUGCG ............((((((.((.(((..........)))..)).))((((((((((.(((((.......)))))........))))))))))(((((((((....)))))))))..)))). ( -45.90) >DroSec_CAF1 861 120 - 1 AUUUUCACGAAUGCGCGUGUAAAGCGAAAAUCGAGGGUGGUGGACGUUGCCAGAUAGCGUGCGAAAAACACGUAACCAACGAUCUGGCAAUGCCGCGCGUUUAUGCGGGUGGCGAGUGUG .....(((...(((.(((((((((((.....).......((((.(((((((((((.(((((.......)))))........))))))))))))))))).)))))))).)))....))).. ( -41.80) >DroSim_CAF1 484 120 - 1 AUUUUCACGAAUGCGCGUGUAAAGCGAAAAUCGAGGGUGGUGGACGUUGCCAGAUAGCGAGCGAAAAACACGUAACCAACGAUCUGGCAAUGCCGCGCGUUUAUGCGGGUGGCGACUGUG .......((...((.(((((((((((.....).......((((.(((((((((((.(....)........(((.....)))))))))))))))))))).)))))))).))..))...... ( -37.50) >DroEre_CAF1 1265 119 - 1 AUUUUCACGAAUGCGCGUGCAAAGCGAAAAUCGAGGGAGGCGGGCGUUGCCAGAUACCGUGCGAAAAACACGUAACCAAC-AUCUGGCAAUGCUGCGCGUUUAUGCGGGAGGCGAGUGCG .....(((((((((((.(((....((.....))......)))(((((((((((((....((((.......))))......-))))))))))))))))))))).(((.....))).))).. ( -41.80) >DroYak_CAF1 457 120 - 1 AUUUUCACGAAUGCGCGUGCAAAGCGAAAAUCGAGAGAGGCGAGCGUUGCCAGAUAUCUUGCGAAAAACACGUAACCAACGACCUGGCAAUGCUGCGCAUUUAUGCGGGUGGAGAGUGCU .....(((...(.(((.((((.........((....)).((((((((((((((.....(((((.......)))))........))))))))))).))).....)))).))).)..))).. ( -39.72) >consensus AUUUUCACGAAUGCGCGUGCAAAGCGAAAAUCGAGGGUGGUGGACGUUGCCAGAUAGCGUGCGAAAAACACGUAACCAACGAUCUGGCAAUGCCGCGCGUUUAUGCGGGUGGCGAGUGCG .....(((..((.((((((.....((.....))......((((.(((((((((((....((((.......)))).......))))))))))))))).....)))))).)).....))).. (-32.78 = -33.30 + 0.52)


| Location | 16,079,003 – 16,079,120 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.20 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -24.91 |
| Energy contribution | -25.43 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16079003 117 - 23771897 ACAAAAAAUUUGGCAGCGACGGU---GCCAGAACGCCGCCAUUUUCACGAAUGCGCGUGCAAAGCGAAAAUCGAGGCUGGUGGACGUUGCCAGAUAGCGUGCGAAAAACACGUAACCAAC .......(((((((((((.((((---(......)))))(((((.((.(((...(((.......)))....))).))..))))).))))))))))).(((((.......)))))....... ( -39.50) >DroPse_CAF1 4189 108 - 1 ACAAAAAAUUUGGCAGCGACAGUAGUUCGACAACGCCGCCAUUUUCACGAAUGCGCGUGCAAAA---UAAUCGUCUGGAGC-GGCGUUGCCAGA-AGCUGGCGGAAAAUUGUU------- ((((....((((.(((((((....))).(.(((((((((.(((((((((......)))).))))---)..((.....))))-))))))).)...-.)))).))))...)))).------- ( -32.80) >DroSim_CAF1 524 117 - 1 ACAAAAAAUUUGGCAGCGACGGU---GCCAGAACGCCGCCAUUUUCACGAAUGCGCGUGUAAAGCGAAAAUCGAGGGUGGUGGACGUUGCCAGAUAGCGAGCGAAAAACACGUAACCAAC .......(((((((((((...((---......)).(((((((((((.......(((.......)))......))))))))))).))))))))))).....(((.......)))....... ( -36.62) >DroEre_CAF1 1304 117 - 1 ACAAAAAAUUUGGCAGCGACGGU---GCCAGAACGCCGCCAUUUUCACGAAUGCGCGUGCAAAGCGAAAAUCGAGGGAGGCGGGCGUUGCCAGAUACCGUGCGAAAAACACGUAACCAAC .......((((((((((...(((---(......))))(((.((..(.(((...(((.......)))....))).)..))...)))))))))))))..((((.......))))........ ( -38.40) >DroYak_CAF1 497 117 - 1 ACAAAAAAUUUGGCAGCGACGGU---GCCAGAACGCCGCCAUUUUCACGAAUGCGCGUGCAAAGCGAAAAUCGAGAGAGGCGAGCGUUGCCAGAUAUCUUGCGAAAAACACGUAACCAAC .......(((((((((((.((((---(......))))(((..((((.(((...(((.......)))....))).)))))))..))))))))))))...(((((.......)))))..... ( -37.70) >DroPer_CAF1 3114 108 - 1 ACAAAAAAUUUGGCAGCGACAGUAGUUCGACAACGCCGCCAUUUUCACGAAUGCGCGUGCAAAA---UAAUCGUCUGGAGC-GGCGUUGCCAGA-AGCUGGCGGAAAAUUGUU------- ((((....((((.(((((((....))).(.(((((((((.(((((((((......)))).))))---)..((.....))))-))))))).)...-.)))).))))...)))).------- ( -32.80) >consensus ACAAAAAAUUUGGCAGCGACGGU___GCCAGAACGCCGCCAUUUUCACGAAUGCGCGUGCAAAGCGAAAAUCGAGGGAGGCGGGCGUUGCCAGAUAGCGUGCGAAAAACACGUAACCAAC .......(((((((((((..((.....)).....((((((((((....))))).))).))........................)))))))))))......................... (-24.91 = -25.43 + 0.53)


| Location | 16,079,120 – 16,079,233 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 95.18 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -24.56 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16079120 113 + 23771897 ACAUCAAAAUAAUCAAAAUAAAGCAUUCUUCCAGUGCUCAACAAUCAGUCCGUGCGAGCACGCGCGAGUGUGUGUGUGCGCAGGAAAACCCGCCGAUCGGGA-AAAGUGUAGA-A ......................(((((.....)))))((.(((.....((((((((.((((((....))))))...))))).)))...((((.....)))).-....))).))-. ( -32.20) >DroSec_CAF1 1018 112 + 1 ACAUCAAAAUAAUCAAAAUAAAGCAUUCUUCCAGUGCUCAAAAAUCAGUCCGUGCGAGCACGCGCGUGUGUGUGUGUGCGCAGGA-AACCCGCCGAUCGGGA-AAAGUGUAGA-A ((((.................((((((.....))))))..........(((.((((.((((((((....)))))))).)))))))-..((((.....)))).-...))))...-. ( -31.70) >DroSim_CAF1 641 113 + 1 ACAUCAAAAUAAUCAAAAUAAAGCAUUCUUCCAGUGCUCAACAAUCAGUCCGUGCGAGCACGCGCGUGUGUGUGUGUGCGCAGGAAAACCCGCCGAUCGGGA-AAAGUGUAGA-A ......................(((((.....)))))((.(((.....(((.((((.((((((((....)))))))).)))))))...((((.....)))).-....))).))-. ( -32.20) >DroEre_CAF1 1421 114 + 1 ACAUCAAAAUAAUCAAAAUAAAGCAUUCUUCCAGUGCUCAACAAUCAGUCCGUGCGAGCACGCGGGUGUGUGUGUGUGCGCAGGAAAAGCCGCCGAUCGGGA-AAAGUGUAGAAA ......................(((((.((((.((((((.((.........))..))))))((((.(.(.((((....)))).)...).))))......)))-).)))))..... ( -31.40) >DroYak_CAF1 614 112 + 1 ACAUCAAAAUAAUCAAAAUAAAGCAUUCUUCCAGUGCUCAACAAUCAGUCCGUUCGAGCACGCGGGUGUGU--GUGUGCGCAGGAAAA-CCGCCGAUCGGAAAAAAGUGUAAAAA ......................(((((.((((.(((((((((.........))).))))))((((.(.(((--(....)))).)....-)))).....))))...)))))..... ( -29.00) >consensus ACAUCAAAAUAAUCAAAAUAAAGCAUUCUUCCAGUGCUCAACAAUCAGUCCGUGCGAGCACGCGCGUGUGUGUGUGUGCGCAGGAAAACCCGCCGAUCGGGA_AAAGUGUAGA_A ((((.................((((((.....))))))..........((((..((.(((((((((....)))))))))((.((.....))))))..)))).....))))..... (-24.56 = -25.00 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:33 2006