
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,077,491 – 16,077,610 |
| Length | 119 |
| Max. P | 0.655940 |

| Location | 16,077,491 – 16,077,610 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.53 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -15.39 |
| Energy contribution | -12.67 |
| Covariance contribution | -2.71 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16077491 119 - 23771897 UUUUAUUUAACCGAUUUCAGGUAAUUGCCAUUUGCAUUAACUGUUUUUUAACCAUGCACAACUAGGGCUAA-AAUGCCGUUGGAAAAGAGAUCGCCACGUGUGCUCACUCGCAUGUGGGC ...........(((((((.(((....)))...(((((.....(((....))).)))))((((...(((...-...))))))).....)))))))(((((((((......))))))))).. ( -33.00) >DroPse_CAF1 5305 118 - 1 UUUC-ACCAAGCGAUUUGAGGUAAUUGAAAUGA-CGAAAAUUGUGUUUAAAUCAUGUGAAAAAUGUGUUACCUGCACAGUUUCACAUUCAAUCCACAUGUGUGGAUGCUUGCAUGUCAGU ....-..((((((((((((..(((((.......-....)))))...)))))))((((((((..(((((.....))))).))))))))...((((((....)))))))))))......... ( -30.80) >DroSec_CAF1 6306 119 - 1 UUUUAUUUAACCGAUUUCAGGUGAUUGCCAUUUGUAUUAACUGUAAUUUAGCCAUUCGCAACUAGAGAUAA-AGUGACGUUGGAAAAGAGAUCGCCACGUGUGCUUACUCGCAUGUGGGC ...........(((((((.(((....)))......................(((..((..(((........-)))..)).)))....)))))))(((((((((......))))))))).. ( -29.50) >DroSim_CAF1 12464 119 - 1 UUUUAUUUAACCGAUUUCAGGUAAUUGCCAUUUGUAUUAACUGUUAUUUAGCCAUGCGCAACUAGGGAUAA-AGUGCCGUUGGAAAAGAGAUCGCCACGUGUGCUUACUCGCAUGUGGGC ...........(((((((.(((....)))......................(((((.(((.((........-))))))).)))....)))))))(((((((((......))))))))).. ( -29.90) >DroYak_CAF1 14479 119 - 1 UUUUAUUUAGCAGAAUUGAGGUAAUUAUAAUUUCUAUUAACUGUAAUUUAGCCAUGCGCAACGUGAGCUAA-AGUGCCGUUGAAAAAGAGUUCGCCACGUGUGUUCACUCGCAUGUGGAC ............((((((((((.......)))))..(((((.(((.((((((((((.....)))).)))))-).))).))))).....))))).(((((((((......))))))))).. ( -33.60) >DroPer_CAF1 5309 107 - 1 ------------GAUUUGAGGUAAUUGAAAUGA-CGAAAAUUGUGUUUAAAUCAUGUGAAAAAUGUGUUACCUGCACAGUUUCACAUCCAAUCCACAUGUGUGGAUGCUUGCAUGUCAGU ------------...........(((((.(((.-(((................((((((((..(((((.....))))).))))))))...((((((....))))))..)))))).))))) ( -26.20) >consensus UUUUAUUUAACCGAUUUCAGGUAAUUGCAAUUUGCAUUAACUGUAAUUUAACCAUGCGCAACUAGAGAUAA_AGUGCCGUUGGAAAAGAGAUCGCCACGUGUGCUUACUCGCAUGUGGGC ............((((((.(((...((.......))...)))....((((((...(((................))).))))))...)))))).(((((((((......))))))))).. (-15.39 = -12.67 + -2.71)

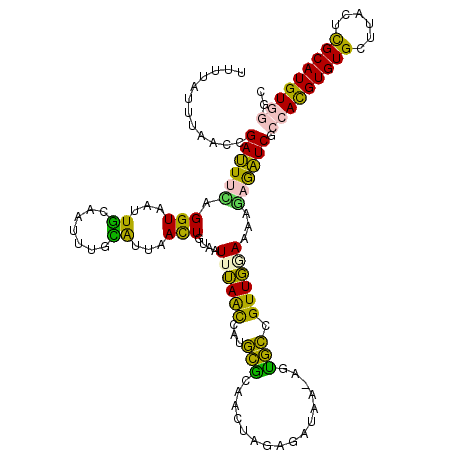
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:28 2006